In Silico Serotyping Based on Whole-Genome Sequencing Improves the Accuracy of Shigella Identification
- PMID: 30709819
- PMCID: PMC6585509
- DOI: 10.1128/AEM.00165-19
In Silico Serotyping Based on Whole-Genome Sequencing Improves the Accuracy of Shigella Identification
Abstract
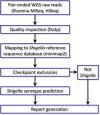
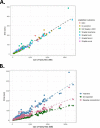
Bacteria of the genus Shigella, consisting of 4 species and >50 serotypes, cause shigellosis, a foodborne disease of significant morbidity, mortality, and economic loss worldwide. Classical Shigella identification based on selective media and serology is tedious, time-consuming, expensive, and not always accurate. A molecular diagnostic assay does not distinguish Shigella at the species level or from enteroinvasive Escherichia coli (EIEC). We inspected genomic sequences from 221 Shigella isolates and observed low concordance rates between conventional designation and molecular serotyping: 86.4% and 80.5% at the species and serotype levels, respectively. Serotype determinants for 6 additional serotypes were identified. Examination of differentiation gene markers commonly perceived as characteristic hallmarks in Shigella showed high variability among different serotypes. Using this information, we developed ShigaTyper, an automated workflow that utilizes limited computational resources to accurately and rapidly determine 59 Shigella serotypes using Illumina paired-end whole-genome sequencing (WGS) reads. Shigella serotype determinants and species-specific diagnostic markers were first identified through read alignment to an in-house curated reference sequence database. Relying on sequence hits that passed a threshold level of coverage and accuracy, serotype could be unambiguously predicted within 1 min for an average-size WGS sample of ∼500 MB. Validation with WGS data from 380 isolates showed an accuracy rate of 98.2%. This pipeline is the first step toward building a comprehensive WGS-based analysis pipeline of Shigella spp. in a field laboratory setting, where speed is essential and resources need to be more cost-effectively dedicated.IMPORTANCEShigella causes diarrheal disease with serious public health implications. However, conventional Shigella identification methods are laborious and time-consuming and can be erroneous due to the high similarity between Shigella and enteroinvasive Escherichia coli (EIEC) and cross-reactivity between serotyping antisera. Further, serotype interpretation is complicated for inexperienced users. To develop an easier method with higher accuracy based on whole-genome sequencing (WGS) for Shigella serotyping, we systematically examined genomic information of Shigella isolates from 53 serotypes to define rules for differentiation and serotyping. We created ShigaTyper, an automated pipeline that accurately and rapidly excludes non-Shigella isolates and identifies 59 Shigella serotypes using Illumina paired-end WGS reads. A serotype can be unambiguously predicted at a data processing speed of 538 MB/min with 98.2% accuracy from a regular laptop. Once it is installed, training in bioinformatics analysis and Shigella genetics is not required. This pipeline is particularly useful to general microbiologists in field laboratories.
Keywords: Shigella; in silico; serotying; whole-genome sequencing.
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Figures


Similar articles
-
Evaluation of a Culture-Dependent Algorithm and a Molecular Algorithm for Identification of Shigella spp., Escherichia coli, and Enteroinvasive E. coli.J Clin Microbiol. 2018 Sep 25;56(10):e00510-18. doi: 10.1128/JCM.00510-18. Print 2018 Oct. J Clin Microbiol. 2018. PMID: 30021824 Free PMC article.
-
Cluster-specific gene markers enhance Shigella and enteroinvasive Escherichia coli in silico serotyping.Microb Genom. 2021 Dec;7(12):000704. doi: 10.1099/mgen.0.000704. Microb Genom. 2021. PMID: 34889728 Free PMC article.
-
Whole genome sequencing of Streptococcus pneumoniae: development, evaluation and verification of targets for serogroup and serotype prediction using an automated pipeline.PeerJ. 2016 Sep 14;4:e2477. doi: 10.7717/peerj.2477. eCollection 2016. PeerJ. 2016. PMID: 27672516 Free PMC article.
-
Historical, current, and emerging tools for identification and serotyping of Shigella.Braz J Microbiol. 2021 Dec;52(4):2043-2055. doi: 10.1007/s42770-021-00573-5. Epub 2021 Sep 15. Braz J Microbiol. 2021. PMID: 34524650 Free PMC article. Review.
-
Relationship among Shigella spp. and enteroinvasive Escherichia coli (EIEC) and their differentiation.Braz J Microbiol. 2015 Mar 4;45(4):1131-8. doi: 10.1590/s1517-83822014000400002. eCollection 2014. Braz J Microbiol. 2015. PMID: 25763015 Free PMC article. Review.
Cited by
-
Towards a Four-Component GMMA-Based Vaccine against Shigella.Vaccines (Basel). 2022 Feb 18;10(2):328. doi: 10.3390/vaccines10020328. Vaccines (Basel). 2022. PMID: 35214786 Free PMC article. Review.
-
Whole-genome sequencing of Shigella for surveillance purposes shows (inter)national relatedness and multidrug resistance in isolates from men who have sex with men.Microb Genom. 2023 Apr;9(4):mgen000978. doi: 10.1099/mgen.0.000978. Microb Genom. 2023. PMID: 37022322 Free PMC article.
-
Characterization of Shigella flexneri in northern Vietnam in 2012-2016.Access Microbiol. 2023 Jun 8;5(6):acmi000493.v4. doi: 10.1099/acmi.0.000493.v4. eCollection 2023. Access Microbiol. 2023. PMID: 37424561 Free PMC article.
-
Whole-genome sequencing for antimicrobial surveillance: species-specific quality thresholds and data evaluation from the network of the European Union Reference Laboratory for Antimicrobial Resistance genomic proficiency tests of 2021 and 2022.mSystems. 2024 Sep 17;9(9):e0016024. doi: 10.1128/msystems.00160-24. Epub 2024 Aug 6. mSystems. 2024. PMID: 39105591 Free PMC article.
-
SeroTools: a Python package for Salmonella serotype data analysis.J Open Source Softw. 2020;5(53):2556. doi: 10.21105/joss.02556. Epub 2020 Sep 5. J Open Source Softw. 2020. PMID: 33817546 Free PMC article. No abstract available.
References
-
- Pires SM, Fischer-Walker CL, Lanata CF, Devleesschauwer B, Hall AJ, Kirk MD, Duarte AS, Black RE, Angulo FJ. 2015. Aetiology-specific estimates of the global and regional incidence and mortality of diarrhoeal diseases commonly transmitted through food. PLoS One 10:e0142927. doi:10.1371/journal.pone.0142927. - DOI - PMC - PubMed
-
- Liu J, Platts-Mills JA, Juma J, Kabir F, Nkeze J, Okoi C, Operario DJ, Uddin J, Ahmed S, Alonso PL, Antonio M, Becker SM, Blackwelder WC, Breiman RF, Faruque AS, Fields B, Gratz J, Haque R, Hossain A, Hossain MJ, Jarju S, Qamar F, Iqbal NT, Kwambana B, Mandomando I, McMurry TL, Ochieng C, Ochieng JB, Ochieng M, Onyango C, Panchalingam S, Kalam A, Aziz F, Qureshi S, Ramamurthy T, Roberts JH, Saha D, Sow SO, Stroup SE, Sur D, Tamboura B, Taniuchi M, Tennant SM, Toema D, Wu Y, Zaidi A, Nataro JP, Kotloff KL, Levine MM, Houpt ER. 2016. Use of quantitative molecular diagnostic methods to identify causes of diarrhoea in children: a reanalysis of the GEMS case-control study. Lancet 388:1291–1301. doi:10.1016/S0140-6736(16)31529-X. - DOI - PMC - PubMed
-
- GBD Diarrhoeal Diseases Collaborators. 2017. Estimates of global, regional, and national morbidity, mortality, and aetiologies of diarrhoeal diseases: a systematic analysis for the Global Burden of Disease Study 2015. Lancet Infect Dis 17:909–948. doi:10.1016/S1473-3099(17)30276-1. - DOI - PMC - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Miscellaneous

