Trait-based community assembly and succession of the infant gut microbiome
- PMID: 30710083
- PMCID: PMC6358638
- DOI: 10.1038/s41467-019-08377-w
Trait-based community assembly and succession of the infant gut microbiome
Abstract
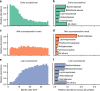
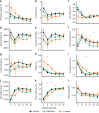
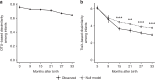
The human gut microbiome develops over early childhood and aids in food digestion and immunomodulation, but the mechanisms driving its development remain elusive. Here we use data curated from literature and online repositories to examine trait-based patterns of gut microbiome succession in 56 infants over their first three years of life. We also develop a new phylogeny-based approach of inferring trait values that can extend readily to other microbial systems and questions. Trait-based patterns suggest that infant gut succession begins with a functionally variable cohort of taxa, adept at proliferating rapidly within hosts, which gradually matures into a more functionally uniform cohort of taxa adapted to thrive in the anoxic gut and disperse between anoxic patches as oxygen-tolerant spores. Trait-based composition stabilizes after the first year, while taxonomic turnover continues unabated, suggesting functional redundancy in the traits examined. Trait-based approaches powerfully complement taxonomy-based approaches to understanding the mechanisms of microbial community assembly and succession.
Conflict of interest statement
The authors declare no competing interests.
Figures


References
-
- Pickett STA, Collins SL, Armesto JJ. Models, mechanisms and pathways of succession. Bot. Rev. 1987;53:335–371. doi: 10.1007/BF02858321. - DOI
-
- Tilman D. Constraints and tradeoffs: toward a predictive theory of competition and succession. Oikos. 1990;58:3. doi: 10.2307/3565355. - DOI
-
- Leibold MA, et al. The metacommunity concept: a framework for multi-scale community ecology. Ecol. Lett. 2004;7:601–613. doi: 10.1111/j.1461-0248.2004.00608.x. - DOI
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources

