Unexpected Receptor Functional Mimicry Elucidates Activation of Coronavirus Fusion
- PMID: 30712865
- PMCID: PMC6751136
- DOI: 10.1016/j.cell.2018.12.028
Unexpected Receptor Functional Mimicry Elucidates Activation of Coronavirus Fusion
Erratum in
-
Unexpected Receptor Functional Mimicry Elucidates Activation of Coronavirus Fusion.Cell. 2020 Dec 10;183(6):1732. doi: 10.1016/j.cell.2020.11.031. Cell. 2020. PMID: 33306956 Free PMC article. No abstract available.
Abstract
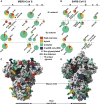
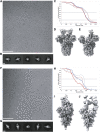
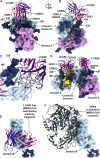
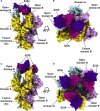
Recent outbreaks of severe acute respiratory syndrome and Middle East respiratory syndrome, along with the threat of a future coronavirus-mediated pandemic, underscore the importance of finding ways to combat these viruses. The trimeric spike transmembrane glycoprotein S mediates entry into host cells and is the major target of neutralizing antibodies. To understand the humoral immune response elicited upon natural infections with coronaviruses, we structurally characterized the SARS-CoV and MERS-CoV S glycoproteins in complex with neutralizing antibodies isolated from human survivors. Although the two antibodies studied blocked attachment to the host cell receptor, only the anti-SARS-CoV S antibody triggered fusogenic conformational changes via receptor functional mimicry. These results provide a structural framework for understanding coronavirus neutralization by human antibodies and shed light on activation of coronavirus membrane fusion, which takes place through a receptor-driven ratcheting mechanism.
Keywords: MERS-CoV; N-linked glycosylation; SARS-CoV; class I fusion protein; coronavirus; glycoproteomics; membrane fusion; neutralizing antibodies; spike glycoprotein.
Copyright © 2018 Elsevier Inc. All rights reserved.
Conflict of interest statement
A.L. is the scientific founder and shareholder of Humabs BioMed. D.C. is currently Chief Scientific Officer of Humabs Biomed. The other authors declare no competing financial interests.
Figures





References
-
- Agirre J., Iglesias-Fernández J., Rovira C., Davies C.J., Wilson K.S., Cowtan K.D. Privateer: software for the conformational validation of carbohydrate structures. Nat. Struct. Mol. Biol. 2015;22:833–834. - PubMed
-
- Blanc E., Roversi P., Vonrhein C., Flensburg C., Lea S.M., Bricogne G. Refinement of severely incomplete structures with maximum likelihood in BUSTER-TNT. Acta Crystallogr. D Biol. Crystallogr. 2004;60:2210–2221. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

