Dynamic Role of the G Protein in Stabilizing the Active State of the Adenosine A2A Receptor
- PMID: 30713025
- PMCID: PMC6531377
- DOI: 10.1016/j.str.2018.12.007
Dynamic Role of the G Protein in Stabilizing the Active State of the Adenosine A2A Receptor
Abstract
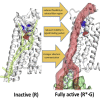
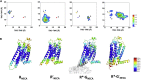
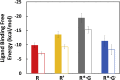
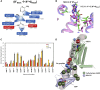
Agonist binding in the extracellular region of the G protein-coupled adenosine A2A receptor increases its affinity to the G proteins in the intracellular region, and vice versa. The structural basis for this effect is not evident from the crystal structures of A2AR in various conformational states since it stems from the receptor dynamics. Using atomistic molecular dynamics simulations on four different conformational states of the adenosine A2A receptor, we observed that the agonists show decreased ligand mobility, lower entropy of the extracellular loops in the active-intermediate state compared with the inactive state. In contrast, the entropy of the intracellular region increases to prime the receptor for coupling the G protein. Coupling of the G protein to A2AR shrinks the agonist binding site, making tighter receptor agonist contacts with an increase in the strength of allosteric communication compared with the active-intermediate state. These insights provide a strong basis for structure-based ligand design studies.
Keywords: G protein-coupled receptors (GPCRs); adenosine receptor; allosteric communication; entropy; ligand mobility; molecular dynamics simulations.
Copyright © 2018. Published by Elsevier Ltd.
Figures




Similar articles
-
Mechanistic insights into allosteric regulation of the A2A adenosine G protein-coupled receptor by physiological cations.Nat Commun. 2018 Apr 10;9(1):1372. doi: 10.1038/s41467-018-03314-9. Nat Commun. 2018. PMID: 29636462 Free PMC article.
-
Agonist-bound adenosine A2A receptor structures reveal common features of GPCR activation.Nature. 2011 May 18;474(7352):521-5. doi: 10.1038/nature10136. Nature. 2011. PMID: 21593763 Free PMC article.
-
Structural and energetic effects of A2A adenosine receptor mutations on agonist and antagonist binding.PLoS One. 2014 Oct 6;9(10):e108492. doi: 10.1371/journal.pone.0108492. eCollection 2014. PLoS One. 2014. PMID: 25285959 Free PMC article.
-
Novel approaches for targeting the adenosine A2A receptor.Expert Opin Drug Discov. 2015 Jan;10(1):63-80. doi: 10.1517/17460441.2015.971006. Epub 2014 Oct 14. Expert Opin Drug Discov. 2015. PMID: 25311639 Review.
-
Allosteric mechanisms within the adenosine A2A-dopamine D2 receptor heterotetramer.Neuropharmacology. 2016 May;104:154-60. doi: 10.1016/j.neuropharm.2015.05.028. Epub 2015 Jun 4. Neuropharmacology. 2016. PMID: 26051403 Free PMC article. Review.
Cited by
-
Allosteric communication regulates ligand-specific GPCR activity.FEBS J. 2021 Apr;288(8):2502-2512. doi: 10.1111/febs.15826. Epub 2021 Apr 5. FEBS J. 2021. PMID: 33738925 Free PMC article. Review.
-
Molecular Simulations and Drug Discovery of Adenosine Receptors.Molecules. 2022 Mar 22;27(7):2054. doi: 10.3390/molecules27072054. Molecules. 2022. PMID: 35408454 Free PMC article. Review.
-
The atomistic level structure for the activated human κ-opioid receptor bound to the full Gi protein and the MP1104 agonist.Proc Natl Acad Sci U S A. 2020 Mar 17;117(11):5836-5843. doi: 10.1073/pnas.1910006117. Epub 2020 Mar 3. Proc Natl Acad Sci U S A. 2020. PMID: 32127473 Free PMC article.
-
Comparative Saturation Binding Analysis of 64Cu-Labeled Somatostatin Analogues Using Cell Homogenates and Intact Cells.ACS Omega. 2023 Jun 22;8(26):24003-24009. doi: 10.1021/acsomega.3c02755. eCollection 2023 Jul 4. ACS Omega. 2023. PMID: 37426243 Free PMC article.
-
Conformational plasticity of the intracellular cavity of GPCR-G-protein complexes leads to G-protein promiscuity and selectivity.Proc Natl Acad Sci U S A. 2019 Jun 11;116(24):11956-11965. doi: 10.1073/pnas.1820944116. Epub 2019 May 28. Proc Natl Acad Sci U S A. 2019. PMID: 31138704 Free PMC article.
References
-
- Berendsen H.J.C., Grigera J.R., Straatsma T.P. The missing term in effective pair potentials. J. Phys. Chem. 1987;91:6269–6271.
-
- Cheng R.K.Y., Segala E., Robertson N., Deflorian F., Dore A.S., Errey J.C., Fiez-Vandal C., Marshall F.H., Cooke R.M. Structures of human A1 and A2A adenosine receptors with xanthines reveal determinants of selectivity. Structure. 2017;25:1275–1285.e4. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

