Molecular methods for assessment of non-covalent metallodrug-DNA interactions
- PMID: 30714595
- PMCID: PMC6657641
- DOI: 10.1039/c8cs00157j
Molecular methods for assessment of non-covalent metallodrug-DNA interactions
Abstract
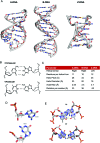
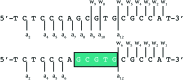
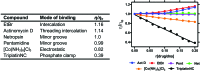
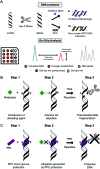
The binding of small molecule metallodrugs to discrete regions of nucleic acids is an important branch of medicinal chemistry and the nature of these interactions, allied with sequence selectivity, forms part of the backbone of modern medicinal inorganic chemistry research. In this tutorial review we describe a range of molecular methods currently employed within our laboratories to explore novel metallodrug-DNA interactions. At the outset, an introduction to DNA from a structural perspective is provided along with descriptions of non-covalent DNA recognition focusing on intercalation, insertion, and phosphate binding. Molecular methods, described from a non-expert perspective, to identify non-covalent and pre-associative nucleic acid recognition are then demonstrated using a variety of techniques including direct (non-optical) and indirect (optical) methods. Direct methods include: X-ray crystallography; NMR spectroscopy; mass spectrometry; and viscosity while indirect approaches detail: competitive inhibition experiments; fluorescence and absorbance spectroscopy; circular dichroism; and electrophoresis-based techniques. For each method described we provide an overview of the technique, a detailed examination of results obtained and relevant follow-on of advanced biophysical/analytical techniques. To achieve this, a selection of relevant copper(ii) and platinum(ii) complexes developed within our laboratories are discussed and are compared, where possible, to classical DNA binding agents. Applying these molecular methods enables us to determine structure-activity factors important to rational metallodrug design. In many cases, combinations of molecular methods are required to comprehensively elucidate new metallodrug-DNA interactions and, from a drug discovery perspective, coupling this data with cellular responses helps to inform understanding of how metallodrug-DNA binding interactions manifest cytotoxic action.
Figures








References
-
- Barry N. P. E., Sadler P. J. Chem. Commun. 2013;49:5106–5131. - PubMed
-
- Chargaff E. Science. 1971;172:637–642. - PubMed
-
- Wing R., Drew H., Takano T., Broka C., Tanaka S., Itakura K., Dickerson R. E. Nature. 1980;287:755–758. - PubMed
-
- Rich A., Zhang S. Nat. Rev. Genet. 2003;4:566–572. - PubMed
-
- Drew H. R., Dickerson R. E. J. Mol. Biol. 1981;152:723–736. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

