Pan-genome analyses of model fungal species
- PMID: 30714895
- PMCID: PMC6421352
- DOI: 10.1099/mgen.0.000243
Pan-genome analyses of model fungal species
Abstract
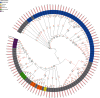
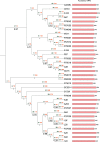
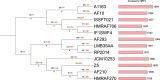
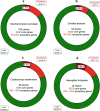
The concept of the species 'pan-genome', the union of 'core' conserved genes and all 'accessory' non-conserved genes across all strains of a species, was first proposed in prokaryotes to account for intraspecific variability. Species pan-genomes have been extensively studied in prokaryotes, but evidence of species pan-genomes has also been demonstrated in eukaryotes such as plants and fungi. Using a previously published methodology based on sequence homology and conserved microsynteny, in addition to bespoke pipelines, we have investigated the pan-genomes of four model fungal species: Saccharomyces cerevisiae, Candida albicans, Cryptococcus neoformans var. grubii and Aspergillus fumigatus. Between 80 and 90 % of gene models per strain in each of these species are core genes that are highly conserved across all strains of that species, many of which are involved in housekeeping and conserved survival processes. In many of these species, the remaining 'accessory' gene models are clustered within subterminal regions and may be involved in pathogenesis and antimicrobial resistance. Analysis of the ancestry of species core and accessory genomes suggests that fungal pan-genomes evolve by strain-level innovations such as gene duplication as opposed to wide-scale horizontal gene transfer. Our findings lend further supporting evidence to the existence of species pan-genomes in eukaryote taxa.
Keywords: Aspergillus; Cryptococcus; comparative genomics; fungal pangenomes; yeast.
Conflict of interest statement
The authors declare that there are no conflicts of interest.
Figures


References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

