Genetic redundancy fuels polygenic adaptation in Drosophila
- PMID: 30716062
- PMCID: PMC6375663
- DOI: 10.1371/journal.pbio.3000128
Genetic redundancy fuels polygenic adaptation in Drosophila
Abstract
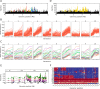
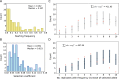
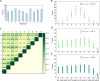
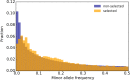
The genetic architecture of adaptive traits is of key importance to predict evolutionary responses. Most adaptive traits are polygenic-i.e., result from selection on a large number of genetic loci-but most molecularly characterized traits have a simple genetic basis. This discrepancy is best explained by the difficulty in detecting small allele frequency changes (AFCs) across many contributing loci. To resolve this, we use laboratory natural selection to detect signatures for selective sweeps and polygenic adaptation. We exposed 10 replicates of a Drosophila simulans population to a new temperature regime and uncovered a polygenic architecture of an adaptive trait with high genetic redundancy among beneficial alleles. We observed convergent responses for several phenotypes-e.g., fitness, metabolic rate, and fat content-and a strong polygenic response (99 selected alleles; mean s = 0.059). However, each of these selected alleles increased in frequency only in a subset of the evolving replicates. We discerned different evolutionary paradigms based on the heterogeneous genomic patterns among replicates. Redundancy and quantitative trait (QT) paradigms fitted the experimental data better than simulations assuming independent selective sweeps. Our results show that natural D. simulans populations harbor a vast reservoir of adaptive variation facilitating rapid evolutionary responses using multiple alternative genetic pathways converging at a new phenotypic optimum. This key property of beneficial alleles requires the modification of testing strategies in natural populations beyond the search for convergence on the molecular level.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures


References
-
- Jones MR, Milles LS, Alves PC, Callahan CM, Alves JM, Lafferty DJR, et al. Adaptive introgression underlies polymorphic seasonal camouflage in snowshoe hares. Science. 2018;1358:1355–8. - PubMed
Publication types
MeSH terms
Associated data
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

