A census-based estimate of Earth's bacterial and archaeal diversity
- PMID: 30716065
- PMCID: PMC6361415
- DOI: 10.1371/journal.pbio.3000106
A census-based estimate of Earth's bacterial and archaeal diversity
Abstract
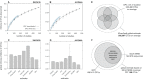
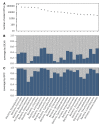
The global diversity of Bacteria and Archaea, the most ancient and most widespread forms of life on Earth, is a subject of intense controversy. This controversy stems largely from the fact that existing estimates are entirely based on theoretical models or extrapolations from small and biased data sets. Here, in an attempt to census the bulk of Earth's bacterial and archaeal ("prokaryotic") clades and to estimate their overall global richness, we analyzed over 1.7 billion 16S ribosomal RNA amplicon sequences in the V4 hypervariable region obtained from 492 studies worldwide, covering a multitude of environments and using multiple alternative primers. From this data set, we recovered 739,880 prokaryotic operational taxonomic units (OTUs, 16S-V4 gene clusters at 97% similarity), a commonly used measure of microbial richness. Using several statistical approaches, we estimate that there exist globally about 0.8-1.6 million prokaryotic OTUs, of which we recovered somewhere between 47%-96%, representing >99.98% of prokaryotic cells. Consistent with this conclusion, our data set independently "recaptured" 91%-93% of 16S sequences from multiple previous global surveys, including PCR-independent metagenomic surveys. The distribution of relative OTU abundances is consistent with a log-normal model commonly observed in larger organisms; the total number of OTUs predicted by this model is also consistent with our global richness estimates. By combining our estimates with the ratio of full-length versus partial-length (V4) sequence diversity in the SILVA sequence database, we further estimate that there exist about 2.2-4.3 million full-length OTUs worldwide. When restricting our analysis to the Americas, while controlling for the number of studies, we obtain similar richness estimates as for the global data set, suggesting that most OTUs are globally distributed. Qualitatively similar results are also obtained for other 16S similarity thresholds (90%, 95%, and 99%). Our estimates constrain the extent of a poorly quantified rare microbial biosphere and refute recent predictions that there exist trillions of prokaryotic OTUs.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures

Comment in
-
Vast (but avoidable) underestimation of global biodiversity.PLoS Biol. 2021 Aug 12;19(8):e3001192. doi: 10.1371/journal.pbio.3001192. eCollection 2021 Aug. PLoS Biol. 2021. PMID: 34383760 Free PMC article.
-
Response to "Vast (but avoidable) underestimation of global biodiversity".PLoS Biol. 2021 Aug 13;19(8):e3001362. doi: 10.1371/journal.pbio.3001362. eCollection 2021 Aug. PLoS Biol. 2021. PMID: 34388158 Free PMC article.
References
-
- Dykhuizen DE. Santa Rosalia revisited: why are there so many species of bacteria? Antonie Leeuwenhoek. 1998; 73(1):25–33. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

