Molecular insights in the pathogenesis of classical Ehlers-Danlos syndrome from transcriptome-wide expression profiling of patients' skin fibroblasts
- PMID: 30716086
- PMCID: PMC6361458
- DOI: 10.1371/journal.pone.0211647
Molecular insights in the pathogenesis of classical Ehlers-Danlos syndrome from transcriptome-wide expression profiling of patients' skin fibroblasts
Abstract
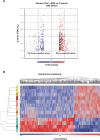
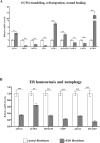
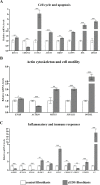
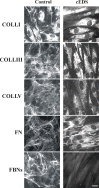
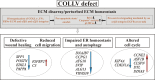
Classical Ehlers-Danlos syndrome (cEDS) is a dominant inherited connective tissue disorder mainly caused by mutations in the COL5A1 and COL5A2 genes encoding type V collagen (COLLV), which is a fibrillar COLL widely distributed in a variety of connective tissues. cEDS patients suffer from skin hyperextensibility, abnormal wound healing/atrophic scars, and joint hypermobility. Most of the causative variants result in a non-functional COL5A1 allele and COLLV haploinsufficiency, whilst COL5A2 mutations affect its structural integrity. To shed light into disease mechanisms involved in cEDS, we performed gene expression profiling in skin fibroblasts from four patients harboring haploinsufficient and structural mutations in both disease genes. Transcriptome profiling revealed significant changes in the expression levels of different extracellular matrix (ECM)-related genes, such as SPP1, POSTN, EDIL3, IGFBP2, and C3, which encode both matricellular and soluble proteins that are mainly involved in cell proliferation and migration, and cutaneous wound healing. These gene expression changes are consistent with our previous protein findings on in vitro fibroblasts from other cEDS patients, which exhibited reduced migration and poor wound repair owing to COLLV disorganization, altered deposition of fibronectin into ECM, and an abnormal integrin pattern. Microarray analysis also indicated the decreased expression of DNAJB7, VIPAS39, CCPG1, ATG10, SVIP, which encode molecular chaperones facilitating protein folding, enzymes regulating post-Golgi COLLs processing, and proteins acting as cargo receptors required for endoplasmic reticulum (ER) proteostasis and implicated in the autophagy process. Patients' cells also showed altered mRNA levels of many cell cycle regulating genes including CCNE2, KIF4A, MKI67, DTL, and DDIAS. Protein studies showed that aberrant COLLV expression causes the disassembly of itself and many structural ECM constituents including COLLI, COLLIII, fibronectin, and fibrillins. Our findings provide the first molecular evidence of significant gene expression changes in cEDS skin fibroblasts highlighting that defective ECM remodeling, ER homeostasis and autophagy might play a role in the pathogenesis of this connective tissue disorder.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures
References
-
- Ritelli M, Dordoni C, Venturini M, Chiarelli N, Quinzani S, Traversa M, et al. Clinical and molecular characterization of 40 patients with classic Ehlers–Danlos syndrome: Identification of 18 COL5A1 and 2 COL5A2 novel mutations. Orphanet J Rare Dis. 2013; 8:58 10.1186/1750-1172-8-58 - DOI - PMC - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

