Identification of a Novel Gene Encoding the Specialized Alanine Decarboxylase in Tea (Camellia sinensis) Plants
- PMID: 30717241
- PMCID: PMC6384637
- DOI: 10.3390/molecules24030540
Identification of a Novel Gene Encoding the Specialized Alanine Decarboxylase in Tea (Camellia sinensis) Plants
Abstract
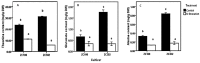
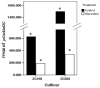
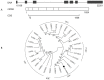
Theanine, a unique amino acid in Camellia sinensis, accounts for more than 50% of total free amino acids in tea and has a significant contribution to the quality of green tea. Previous research indicated that theanine is synthesized from glutamic acid (Glu) and ethylamine mainly in roots, and that theanine accumulation depends on the availability of ethylamine which is derived from alanine (Ala) decarboxylation catalyzed by alanine decarboxylase (AlaDC). However, the specific gene encoding AlaDC protein remains to be discovered in tea plants or in other species. To explore the gene of AlaDC in tea plants, the differences in theanine contents and gene expressions between pretreatment and posttreatment of long-time nitrogen starvation were analyzed in young roots of two tea cultivars. A novel gene annotated as serine decarboxylase (SDC) was noted for its expression levels, which showed high consistency with theanine content, and the expression was remarkably high in young roots under sufficient nitrogen condition. To verify its function, full-length complementary DNA (cDNA) of this candidate gene was cloned from young roots of tea seedlings, and the target protein was expressed and purified from Escherichia coli (E. coli). The enzymatic activity of the protein for Ala and Ser was measured in vitro using ultra-performance liquid chromatography coupled with mass spectrometry (UPLC-MS). The results illustrated that the target protein could catalyze the decarboxylation of Ala despite of its high similarity with SDC from other species. Therefore, this novel gene was identified as AlaDC and named CsAlaDC. Furthermore, the gene expression levels of CsAlaDC in different tissues of tea plants were also quantified with quantitative real-time PCR (qRT-PCR). The results suggest that transcription levels of CsAlaDC in root tissues are significantly higher than those in leaf tissues. That may explain why theanine biosynthesis preferentially occurs in the roots of tea plants. The expression of the gene was upregulated when nitrogen was present, suggesting that theanine biosynthesis is regulated by nitrogen supply and closely related to nitrogen metabolism for C. sinensis. The results of this study are significant supplements to the theanine biosynthetic pathway and provide evidence for the differential accumulation of theanine between C. sinensis and other species.
Keywords: Camellia sinensis; alanine decarboxylase; enzymatic activity; ethylamine; gene expression; nitrogen metabolism; theanine.
Conflict of interest statement
The authors have no conflict of interest to declare.
Figures



References
-
- Chen L., Zhou Z.-X., Yang Y.-J. Genetic improvement and breeding of tea plant (Camellia sinensis) in China: From individual selection to hybridization and molecular breeding. Euphytica. 2007;154:239–248. doi: 10.1007/s10681-006-9292-3. - DOI
-
- Zhang H.-B., Xia E.-H., Huang H., Jiang J.-J., Liu B.-Y., Gao L.-Z. De novo transcriptome assembly of the wild relative of tea tree (Camellia taliensis) and comparative analysis with tea transcriptome identified putative genes associated with tea quality and stress response. BMC Genom. 2015;16:298. doi: 10.1186/s12864-015-1494-4. - DOI - PMC - PubMed
-
- Yang Z., Baldermann S., Watanabe N. Recent studies of the volatile compounds in tea. Food Res. Int. 2013;53:585–599. doi: 10.1016/j.foodres.2013.02.011. - DOI
-
- Liang Y.R., Ma W.Y., Lu J.L., Wu Y. Comparison of chemical compositions of Ilex latifolia thumb and Camellia sinensis L. Food Chem. 2001;75:339–343. doi: 10.1016/S0308-8146(01)00209-6. - DOI
-
- Mamati G.E., Liang Y.R., Lu J.L. Expression of basic genes involved in tea polyphenol synthesis in relation to accumulation of catechins and total tea polyphenols. J. Sci. Food Agric. 2006;86:459–464. doi: 10.1002/jsfa.2368. - DOI
MeSH terms
Substances
Grants and funding
- 31570695/National Natural Science Foundation of China
- 2016C02053-6/the Major Science and Technology Special Project of Variety Breeding of Zhejiang Province
- 1610212016016/Central Public-interest Scientific Institution Basal Research Fund
- 1610212018004/Central Public-interest Scientific Institution Basal Research Fund
- CARS-19/Modern Agro-industry Technology Research System
LinkOut - more resources
Full Text Sources

