Single-Cell RNA Sequencing Resolves Molecular Relationships Among Individual Plant Cells
- PMID: 30718350
- PMCID: PMC6446759
- DOI: 10.1104/pp.18.01482
Single-Cell RNA Sequencing Resolves Molecular Relationships Among Individual Plant Cells
Abstract
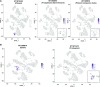
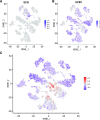
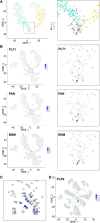
Single-cell RNA sequencing (scRNA-seq) has been used extensively to study cell-specific gene expression in animals, but it has not been widely applied to plants. Here, we describe the use of a commercially available droplet-based microfluidics platform for high-throughput scRNA-seq to obtain single-cell transcriptomes from protoplasts of more than 10,000 Arabidopsis (Arabidopsis thaliana) root cells. We find that all major tissues and developmental stages are represented in this single-cell transcriptome population. Further, distinct subpopulations and rare cell types, including putative quiescent center cells, were identified. A focused analysis of root epidermal cell transcriptomes defined developmental trajectories for individual cells progressing from meristematic through mature stages of root-hair and nonhair cell differentiation. In addition, single-cell transcriptomes were obtained from root epidermis mutants, enabling a comparative analysis of gene expression at single-cell resolution and providing an unprecedented view of the impact of the mutated genes. Overall, this study demonstrates the feasibility and utility of scRNA-seq in plants and provides a first-generation gene expression map of the Arabidopsis root at single-cell resolution.
© 2019 American Society of Plant Biologists. All Rights Reserved.
Figures



Comment in
-
Sequencing Plant Transcriptomes at Single-Cell Resolution Allows Unprecedented Characterization of Genetic and Developmental Cellular Processes.Plant Physiol. 2019 Apr;179(4):1439-1440. doi: 10.1104/pp.19.00199. Plant Physiol. 2019. PMID: 30940738 Free PMC article. No abstract available.
References
-
- Bernhardt C, Zhao M, Gonzalez A, Lloyd A, Schiefelbein J (2005) The bHLH genes GL3 and EGL3 participate in an intercellular regulatory circuit that controls cell patterning in the Arabidopsis root epidermis. Development 132: 291–298 - PubMed
-
- Birnbaum K, Jung JW, Wang JY, Lambert GM, Hirst JA, Galbraith DW, Benfey PN (2005) Cell type-specific expression profiling in plants via cell sorting of protoplasts from fluorescent reporter lines. Nat Methods 2: 615–619 - PubMed
-
- Brady SM, Orlando DA, Lee JY, Wang JY, Koch J, Dinneny JR, Mace D, Ohler U, Benfey PN (2007) A high-resolution root spatiotemporal map reveals dominant expression patterns. Science 318: 801–806 - PubMed
-
- Brennecke P, Anders S, Kim JK, Kołodziejczyk AA, Zhang X, Proserpio V, Baying B, Benes V, Teichmann SA, Marioni JC, Heisler MG (2013) Accounting for technical noise in single-cell RNA-seq experiments. Nat Methods 10: 1093–1095 - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

