Chromosome alignment maintenance requires the MAP RECQL4, mutated in the Rothmund-Thomson syndrome
- PMID: 30718377
- PMCID: PMC6362308
- DOI: 10.26508/lsa.201800120
Chromosome alignment maintenance requires the MAP RECQL4, mutated in the Rothmund-Thomson syndrome
Abstract
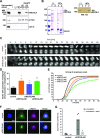
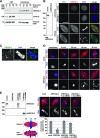
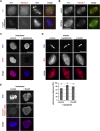
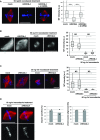
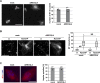
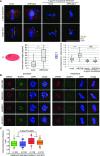
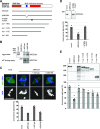
RecQ-like helicase 4 (RECQL4) is mutated in patients suffering from the Rothmund-Thomson syndrome, a genetic disease characterized by premature aging, skeletal malformations, and high cancer susceptibility. Known roles of RECQL4 in DNA replication and repair provide a possible explanation of chromosome instability observed in patient cells. Here, we demonstrate that RECQL4 is a microtubule-associated protein (MAP) localizing to the mitotic spindle. RECQL4 depletion in M-phase-arrested frog egg extracts does not affect spindle assembly per se, but interferes with maintaining chromosome alignment at the metaphase plate. Low doses of nocodazole depolymerize RECQL4-depleted spindles more easily, suggesting abnormal microtubule-kinetochore interaction. Surprisingly, inter-kinetochore distance of sister chromatids is larger in depleted extracts and patient fibroblasts. Consistent with a role to maintain stable chromosome alignment, RECQL4 down-regulation in HeLa cells causes chromosome misalignment and delays mitotic progression. Importantly, these chromosome alignment defects are independent from RECQL4's reported roles in DNA replication and damage repair. Our data elucidate a novel function of RECQL4 in mitosis, and defects in mitotic chromosome alignment might be a contributing factor for the Rothmund-Thomson syndrome.
© 2019 Yokoyama et al.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures





References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials
