Convergent gene losses illuminate metabolic and physiological changes in herbivores and carnivores
- PMID: 30718421
- PMCID: PMC6386725
- DOI: 10.1073/pnas.1818504116
Convergent gene losses illuminate metabolic and physiological changes in herbivores and carnivores
Abstract
The repeated evolution of dietary specialization represents a hallmark of mammalian ecology. To detect genomic changes that are associated with dietary adaptations, we performed a systematic screen for convergent gene losses associated with an obligate herbivorous or carnivorous diet in 31 placental mammals. For herbivores, our screen discovered the repeated loss of the triglyceride lipase inhibitor PNLIPRP1, suggesting enhanced triglyceride digestion efficiency. Furthermore, several herbivores lost the pancreatic exocytosis factor SYCN, providing an explanation for continuous pancreatic zymogen secretion in these species. For carnivores, we discovered the repeated loss of the hormone-receptor pair INSL5-RXFP4 that regulates appetite and glucose homeostasis, which likely relates to irregular feeding patterns and constant gluconeogenesis. Furthermore, reflecting the reduced need to metabolize plant-derived xenobiotics, several carnivores lost the xenobiotic receptors NR1I3 and NR1I2 Finally, the carnivore-associated loss of the gastrointestinal host defense gene NOX1 could be related to a reduced gut microbiome diversity. By revealing convergent gene losses associated with differences in dietary composition, feeding patterns, and gut microbiomes, our study contributes to understanding how similar dietary specializations evolved repeatedly in mammals.
Keywords: carnivorous diet; convergent gene loss; herbivorous diet; metabolism; physiology.
Copyright © 2019 the Author(s). Published by PNAS.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
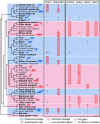


References
-
- Mackie RI. Mutualistic fermentative digestion in the gastrointestinal tract: Diversity and evolution. Integr Comp Biol. 2002;42:319–326. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

