A cancer-associated point mutation disables the steric gate of human PrimPol
- PMID: 30718533
- PMCID: PMC6362072
- DOI: 10.1038/s41598-018-37439-0
A cancer-associated point mutation disables the steric gate of human PrimPol
Abstract
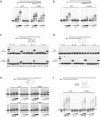
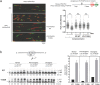
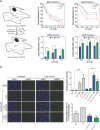
PrimPol is a human primase/polymerase specialized in re-starting stalled forks by repriming beyond lesions such as pyrimidine dimers, and replication-perturbing structures including G-quadruplexes and R-loops. Unlike most conventional primases, PrimPol proficiently discriminates against ribonucleotides (NTPs), being able to start synthesis using deoxynucleotides (dNTPs), yet the structural basis and physiological implications for this discrimination are not understood. In silico analyses based on the three-dimensional structure of human PrimPol and related enzymes enabled us to predict a single residue, Tyr100, as the main effector of sugar discrimination in human PrimPol and a change of Tyr100 to histidine to boost the efficiency of NTP incorporation. We show here that the Y100H mutation profoundly stimulates NTP incorporation by human PrimPol, with an efficiency similar to that for dNTP incorporation during both primase and polymerase reactions in vitro. As expected from the higher cellular concentration of NTPs relative to dNTPs, Y100H expression in mouse embryonic fibroblasts and U2OS osteosarcoma cells caused enhanced resistance to hydroxyurea, which decreases the dNTP pool levels in S-phase. Remarkably, the Y100H PrimPol mutation has been identified in cancer, suggesting that this mutation could be selected to promote survival at early stages of tumorigenesis, which is characterized by depleted dNTP pools.
Conflict of interest statement
The authors declare no competing interests.
Figures


References
-
- Li Y, Breaker RR. Kinetics of RNA degradation by specific base catalysis of transesterification involving the 2′-hydroxyl group. J Am Chem Soc. 1999;121:5364–5372. doi: 10.1021/ja990592p. - DOI
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

