Mycobacterium tuberculosis Rv0366c-Rv0367c encodes a non-canonical PezAT-like toxin-antitoxin pair
- PMID: 30718534
- PMCID: PMC6362051
- DOI: 10.1038/s41598-018-37473-y
Mycobacterium tuberculosis Rv0366c-Rv0367c encodes a non-canonical PezAT-like toxin-antitoxin pair
Abstract
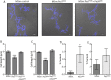
Toxin-antitoxin (TA) systems are ubiquitously existing addiction modules with essential roles in bacterial persistence and virulence. The genome of Mycobacterium tuberculosis encodes approximately 79 TA systems. Through computational and experimental investigations, we report for the first time that Rv0366c-Rv0367c is a non-canonical PezAT-like toxin-antitoxin system in M. tuberculosis. Homology searches with known PezT homologues revealed that residues implicated in nucleotide, antitoxin-binding and catalysis are conserved in Rv0366c. Unlike canonical PezA antitoxins, the N-terminal of Rv0367c is predicted to adopt the ribbon-helix-helix (RHH) motif for deoxyribonucleic acid (DNA) recognition. Further, the modelled complex predicts that the interactions between PezT and PezA involve conserved residues. We performed a large-scale search in sequences encoded in 101 mycobacterial and 4500 prokaryotic genomes and show that such an atypical PezAT organization is conserved in 20 other mycobacterial organisms and in families of class Actinobacteria. We also demonstrate that overexpression of Rv0366c induces bacteriostasis and this growth defect could be restored upon co-expression of cognate antitoxin, Rv0367c. Further, we also observed that inducible expression of Rv0366c in Mycobacterium smegmatis results in decreased cell-length and enhanced tolerance against a front-line tuberculosis (TB) drug, ethambutol. Taken together, we have identified and functionally characterized a novel non-canonical TA system from M. tuberculosis.
Conflict of interest statement
The authors declare no competing interests.
Figures







References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

