Evolution of substrate-specific gene expression and RNA editing in brown rot wood-decaying fungi
- PMID: 30718807
- PMCID: PMC6776062
- DOI: 10.1038/s41396-019-0359-2
Evolution of substrate-specific gene expression and RNA editing in brown rot wood-decaying fungi
Retraction in
-
Retraction Note: Evolution of substrate-specific gene expression and RNA editing in brown rot wood-decaying fungi.ISME J. 2022 Jan;16(1):322. doi: 10.1038/s41396-021-01156-w. ISME J. 2022. PMID: 34853476 Free PMC article. No abstract available.
Abstract
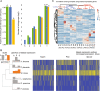
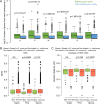
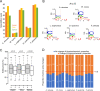
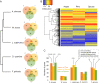
Fungi that decay wood have characteristic associations with certain tree species, but the mechanistic bases for these associations are poorly understood. We studied substrate-specific gene expression and RNA editing in six species of wood-decaying fungi from the 'Antrodia clade' (Polyporales, Agaricomycetes) on three different wood substrates (pine, spruce, and aspen) in submerged cultures. We identified dozens to hundreds of substrate-biased genes (i.e., genes that are significantly upregulated in one substrate relative to the other two substrates) in each species, and these biased genes are correlated with their host ranges. Evolution of substrate-biased genes is associated with gene family expansion, gain and loss of genes, and variation in cis- and trans- regulatory elements, rather than changes in protein coding sequences. We also demonstrated widespread RNA editing events in the Antrodia clade, which differ from those observed in the Ascomycota in their distribution, substitution types, and the genomic environment. Moreover, we found that substrates could affect editing positions and frequency, including editing events occurring in mRNA transcribed from wood-decay-related genes. This work shows the extent to which gene expression and RNA editing differ among species and substrates, and provides clues into mechanisms by which wood-decaying fungi may adapt to different hosts.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures


References
-
- Gilbertson RL. Wood-rotting fungi of North-America. Mycologia. 1980;72:1–49.
-
- Nilsson T, Daniel G, Kirk TK, Obst JR. Chemistry and microscopy of wood decay by some higher ascomyceyes. Holzforschung. 1989;43:11–8.
-
- Wells K, Bandoni RJ. Heterobasidiomycetes. In: Mc Laughlin DJ, Mc Laughlin EG, Lemke PA, editors. The Mycota. Berlin: Springer Verlag; 2001. p. 85–120.
-
- Thiers HD. The secotioid syndrome. Mycologia. 1984;76:1–8.

