A human gut bacterial genome and culture collection for improved metagenomic analyses
- PMID: 30718869
- PMCID: PMC6785715
- DOI: 10.1038/s41587-018-0009-7
A human gut bacterial genome and culture collection for improved metagenomic analyses
Abstract
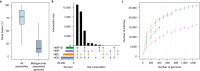
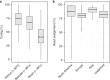
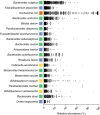
Understanding gut microbiome functions requires cultivated bacteria for experimental validation and reference bacterial genome sequences to interpret metagenome datasets and guide functional analyses. We present the Human Gastrointestinal Bacteria Culture Collection (HBC), a comprehensive set of 737 whole-genome-sequenced bacterial isolates, representing 273 species (105 novel species) from 31 families found in the human gastrointestinal microbiota. The HBC increases the number of bacterial genomes derived from human gastrointestinal microbiota by 37%. The resulting global Human Gastrointestinal Bacteria Genome Collection (HGG) classifies 83% of genera by abundance across 13,490 shotgun-sequenced metagenomic samples, improves taxonomic classification by 61% compared to the Human Microbiome Project (HMP) genome collection and achieves subspecies-level classification for almost 50% of sequences. The improved resource of gastrointestinal bacterial reference sequences circumvents dependence on de novo assembly of metagenomes and enables accurate and cost-effective shotgun metagenomic analyses of human gastrointestinal microbiota.
Conflict of interest statement
S.C.F., B.A.N., M.D., R.D.F. and T.D.L. are either employees of or consultants to Microbiotica Pty Ltd.
Figures


Comment in
-
Human gut bacterial genome reference.Nat Methods. 2019 Apr;16(4):286. doi: 10.1038/s41592-019-0384-0. Nat Methods. 2019. PMID: 30923389 No abstract available.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

