Fusobacterium spp. target human CEACAM1 via the trimeric autotransporter adhesin CbpF
- PMID: 30719234
- PMCID: PMC6346709
- DOI: 10.1080/20002297.2018.1565043
Fusobacterium spp. target human CEACAM1 via the trimeric autotransporter adhesin CbpF
Abstract
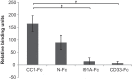
Neisseria meningitidis, Haemophilus influenzae, and Moraxella catarrhalis are pathogenic bacteria adapted to reside on human respiratory mucosal epithelia. One common feature of these species is their ability to target members of the carcinoembryonic antigen-related cell adhesion molecule (CEACAM) family, especially CEACAM1, which is achieved via structurally distinct ligands expressed by each species. Beside respiratory epithelial cells, cells at the dentogingival junction express high levels of CEACAM1. It is possible that bacterial species resident within the oral cavity also utilise CEACAM1 for colonisation and invasion of gingival tissues. From a screen of 59 isolates from the human oral cavity representing 49 bacterial species, we identified strains from Fusobacterium bound to CEACAM1. Of the Fusobacterium species tested, the CEACAM1-binding property was exhibited by F. nucleatum (Fn) and F. vincentii (Fv) but not F. polymorphum (Fp) or F. animalis (Fa) strains tested. These studies identified that CEACAM adhesion was mediated using a trimeric autotransporter adhesin (TAA) for which no function has thus far been defined. We therefore propose the name CEACAM binding protein of Fusobacterium (CbpF). CbpF was identified to be present in the majority of unspeciated Fusobacterium isolates confirming a subset of Fusobacterium spp. are able to target human CEACAM1.
Keywords: CEA; CEACAM1; Fusobacterium; Fusobacterium nucleatum; TAA; Type V secretion; adhesion; binding; host–pathogen interaction; trimeric autotransporter adhesin.
Figures










References
-
- Kim KS.Acute bacterial meningitis in infants and children. Lancet Infect Dis. 2010;10:32–16. - PubMed
-
- Karalus R, Campagnari A. Moraxella catarrhalis: a review of an important human mucosal pathogen. Microbes Infect. 2000;2:547–559. - PubMed
-
- Hill DJ, Virji M. A novel cell-binding mechanism of Moraxella catarrhalis ubiquitous surface protein UspA: specific targeting of the N-domain of carcinoembryonic antigen-related cell adhesion molecules by UspA1. Mol Microbiol. 2003;48:117–129. - PubMed
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous
