Heparin-induced tau filaments are polymorphic and differ from those in Alzheimer's and Pick's diseases
- PMID: 30720432
- PMCID: PMC6375701
- DOI: 10.7554/eLife.43584
Heparin-induced tau filaments are polymorphic and differ from those in Alzheimer's and Pick's diseases
Abstract
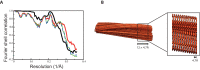
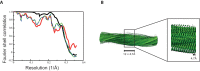
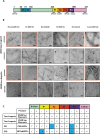
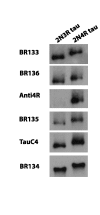
Assembly of microtubule-associated protein tau into filamentous inclusions underlies a range of neurodegenerative diseases. Tau filaments adopt different conformations in Alzheimer's and Pick's diseases. Here, we used cryo- and immuno- electron microscopy to characterise filaments that were assembled from recombinant full-length human tau with four (2N4R) or three (2N3R) microtubule-binding repeats in the presence of heparin. 2N4R tau assembles into multiple types of filaments, and the structures of three types reveal similar 'kinked hairpin' folds, in which the second and third repeats pack against each other. 2N3R tau filaments are structurally homogeneous, and adopt a dimeric core, where the third repeats of two tau molecules pack in a parallel manner. The heparin-induced tau filaments differ from those of Alzheimer's or Pick's disease, which have larger cores with different repeat compositions. Our results illustrate the structural versatility of amyloid filaments, and raise questions about the relevance of in vitro assembly.
Keywords: E.coli over-expression; RELION; cryo-EM; helical reconstruction; human; micro-tubule associated protein tau; molecular biophysics; structural biology.
© 2019, Zhang et al.
Conflict of interest statement
WZ, BF, AM, JF, RC No competing interests declared, MG, SS Reviewing editor, eLife
Figures










References
-
- Adams PD, Afonine PV, Bunkóczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung LW, Kapral GJ, Grosse-Kunstleve RW, McCoy AJ, Moriarty NW, Oeffner R, Read RJ, Richardson DC, Richardson JS, Terwilliger TC, Zwart PH. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallographica Section D Biological Crystallography. 2010;66:213–221. doi: 10.1107/S0907444909052925. - DOI - PMC - PubMed
-
- Andronesi OC, von Bergen M, Biernat J, Seidel K, Griesinger C, Mandelkow E, Baldus M. Characterization of Alzheimer's-like paired helical filaments from the core domain of tau protein using solid-state NMR spectroscopy. Journal of the American Chemical Society. 2008;130:5922–5928. doi: 10.1021/ja7100517. - DOI - PubMed
-
- Annamalai K, Liberta F, Vielberg MT, Close W, Lilie H, Gührs KH, Schierhorn A, Koehler R, Schmidt A, Haupt C, Hegenbart U, Schönland S, Schmidt M, Groll M, Fändrich M. Common fibril structures imply systemically conserved protein misfolding pathways in vivo. Angewandte Chemie International Edition. 2017;56:7510–7514. doi: 10.1002/anie.201701761. - DOI - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- Joint Programme - Neurodegeneration Research REfrAME/European Union/International
- MC_U105192716/MRC_/Medical Research Council/United Kingdom
- MC_UP_A025_1013/MRC_/Medical Research Council/United Kingdom
- IMPRIND-116060/European Union/International
- Joint Programme- Neurodegeneration Research REfrAME/European Union/International
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

