ZincBind-the database of zinc binding sites
- PMID: 30722040
- PMCID: PMC6361820
- DOI: 10.1093/database/baz006
ZincBind-the database of zinc binding sites
Abstract
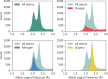
Zinc is one of the most important biologically active metals. Ten per cent of the human genome is thought to encode a zinc binding protein and its uses encompass catalysis, structural stability, gene expression and immunity. At present, there is no specific resource devoted to identifying and presenting all currently known zinc binding sites. Here we present ZincBind, a database of zinc binding sites and its web front-end. Using the structural data in the Protein Data Bank, ZincBind identifies every instance of zinc binding to a protein, identifies its binding site and clusters sites based on 90% sequence identity. There are currently 24 992 binding sites, clustered into 7489 unique sites. The data are available over the web where they can be browsed and downloaded, and via a REST API. ZincBind is regularly updated and will continue to be updated with new data and features.
Figures




References
-
- Prasad A.S., Miale A., Sandstead H.H. et al. (1963) Zinc metabolism in patients with the syndrome of iron deficiency anemia, hepatosplenomegaly, dwarfism, and hypognadism. J. Lab. Clin. Med., 61, 537–549. - PubMed
-
- Liljas A., Kannan K.K., Bergsten P.-C. et al. (1972) Crystal structure of human carbonic anhydrase C. Nat. New Biol., 235:131–137. - PubMed
-
- Bertini I., Luchinat C. and Monnanni R. (1985) Zinc enzymes. J. Chem. Educ., 62, 924.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

