Peptidoglycan Remodeling Enables Escherichia coli To Survive Severe Outer Membrane Assembly Defect
- PMID: 30723128
- PMCID: PMC6428754
- DOI: 10.1128/mBio.02729-18
Peptidoglycan Remodeling Enables Escherichia coli To Survive Severe Outer Membrane Assembly Defect
Abstract
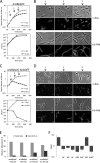
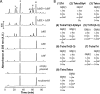
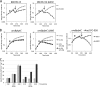
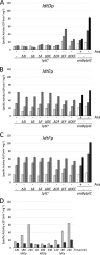
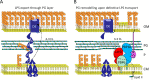
Gram-negative bacteria have a tripartite cell envelope with the cytoplasmic membrane (CM), a stress-bearing peptidoglycan (PG) layer, and the asymmetric outer membrane (OM) containing lipopolysaccharide (LPS) in the outer leaflet. Cells must tightly coordinate the growth of their complex envelope to maintain cellular integrity and OM permeability barrier function. The biogenesis of PG and LPS relies on specialized macromolecular complexes that span the entire envelope. In this work, we show that Escherichia coli cells are capable of avoiding lysis when the transport of LPS to the OM is compromised, by utilizing LD-transpeptidases (LDTs) to generate 3-3 cross-links in the PG. This PG remodeling program relies mainly on the activities of the stress response LDT, LdtD, together with the major PG synthase PBP1B, its cognate activator LpoB, and the carboxypeptidase PBP6a. Our data support a model according to which these proteins cooperate to strengthen the PG in response to defective OM synthesis.IMPORTANCE In Gram-negative bacteria, the outer membrane protects the cell against many toxic molecules, and the peptidoglycan layer provides protection against osmotic challenges, allowing bacterial cells to survive in changing environments. Maintaining cell envelope integrity is therefore a question of life or death for a bacterial cell. Here we show that Escherichia coli cells activate the LD-transpeptidase LdtD to introduce 3-3 cross-links in the peptidoglycan layer when the integrity of the outer membrane is compromised, and this response is required to avoid cell lysis. This peptidoglycan remodeling program is a strategy to increase the overall robustness of the bacterial cell envelope in response to defects in the outer membrane.
Keywords: Escherichia coli; cell envelope; lipopolysaccharide; peptidoglycan; stress response.
Copyright © 2019 Morè et al.
Figures
References
-
- Kamio Y, Nikaido H. 1976. Outer membrane of Salmonella typhimurium: accessibility of phospholipid head groups to phospholipase c and cyanogen bromide activated dextran in the external medium. Biochemistry 15:2561–2570. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
