Targeting NOTCH activation in small cell lung cancer through LSD1 inhibition
- PMID: 30723171
- PMCID: PMC6530478
- DOI: 10.1126/scisignal.aau2922
Targeting NOTCH activation in small cell lung cancer through LSD1 inhibition
Abstract
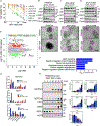
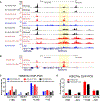
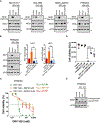
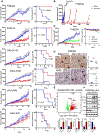
Small cell lung cancer (SCLC) is a recalcitrant, aggressive neuroendocrine-type cancer for which little change to first-line standard-of-care treatment has occurred within the last few decades. Unlike nonsmall cell lung cancer (NSCLC), SCLC harbors few actionable mutations for therapeutic intervention. Lysine-specific histone demethylase 1A (LSD1 also known as KDM1A) inhibitors were previously shown to have selective activity in SCLC models, but the underlying mechanism was elusive. Here, we found that exposure to the selective LSD1 inhibitor ORY-1001 activated the NOTCH pathway, resulting in the suppression of the transcription factor ASCL1 and the repression of SCLC tumorigenesis. Our analyses revealed that LSD1 bound to the NOTCH1 locus, thereby suppressing NOTCH1 expression and downstream signaling. Reactivation of NOTCH signaling with the LSD1 inhibitor reduced the expression of ASCL1 and neuroendocrine cell lineage genes. Knockdown studies confirmed the pharmacological inhibitor-based results. In vivo, sensitivity to LSD1 inhibition in SCLC patient-derived xenograft (PDX) models correlated with the extent of consequential NOTCH pathway activation and repression of a neuroendocrine phenotype. Complete and durable tumor regression occurred with ORY-1001-induced NOTCH activation in a chemoresistant PDX model. Our findings reveal how LSD1 inhibitors function in this tumor and support their potential as a new and targeted therapy for SCLC.
Copyright © 2019 The Authors, some rights reserved; exclusive licensee American Association for the Advancement of Science. No claim to original U.S. Government Works.
Conflict of interest statement
Figures


References
-
- George J, Lim JS, Jang SJ, Cun Y, Ozretic L, Kong G, Leenders F, Lu X, Fernandez-Cuesta L, Bosco G, Muller C, Dahmen I, Jahchan NS, Park KS, Yang D, Karnezis AN, Vaka D, Torres A, Wang MS, Korbel JO, Menon R, Chun SM, Kim D, Wilkerson M, Hayes N, Engelmann D, Putzer B, Bos M, Michels S, Vlasic I, Seidel D, Pinther B, Schaub P, Becker C, Altmuller J, Yokota J, Kohno T, Iwakawa R, Tsuta K, Noguchi M, Muley T, Hoffmann H, Schnabel PA, Petersen I, Chen Y, Soltermann A, Tischler V, Choi CM, Kim YH, Massion PP, Zou Y, Jovanovic D, Kontic M, Wright GM, Russell PA, Solomon B, Koch I, Lindner M, Muscarella LA, la Torre A, Field JK, Jakopovic M, Knezevic J, Castanos-Velez E, Roz L, Pastorino U, Brustugun OT, Lund-Iversen M, Thunnissen E, Kohler J, Schuler M, Botling J, Sandelin M, Sanchez-Cespedes M, Salvesen HB, Achter V, Lang U, Bogus M, Schneider PM, Zander T, Ansen S, Hallek M, Wolf J, Vingron M, Yatabe Y, Travis WD, Nurnberg P, Reinhardt C, Perner S, Heukamp L, Buttner R, Haas SA, Brambilla E, Peifer M, Sage J, Thomas RK, Comprehensive genomic profiles of small cell lung cancer. Nature 524, 47–53 (2015); published online EpubAug 6 (10.1038/nature14664). - DOI - PMC - PubMed
-
- Rudin CM, Durinck S, Stawiski EW, Poirier JT, Modrusan Z, Shames DS, Bergbower EA, Guan Y, Shin J, Guillory J, Rivers CS, Foo CK, Bhatt D, Stinson J, Gnad F, Haverty PM, Gentleman R, Chaudhuri S, Janakiraman V, Jaiswal BS, Parikh C, Yuan W, Zhang Z, Koeppen H, Wu TD, Stern HM, Yauch RL, Huffman KE, Paskulin DD, Illei PB, Varella-Garcia M, Gazdar AF, de Sauvage FJ, Bourgon R, Minna JD, Brock MV, Seshagiri S, Comprehensive genomic analysis identifies SOX2 as a frequently amplified gene in small-cell lung cancer. Nature genetics, (2012); published online EpubSep 2 (10.1038/ng.2405). - DOI - PMC - PubMed
-
- Augert A, Zhang Q, Bates B, Cui M, Wang X, Wildey G, Dowlati A, MacPherson D, Small Cell Lung Cancer Exhibits Frequent Inactivating Mutations in the Histone Methyltransferase KMT2D/MLL2: CALGB 151111 (Alliance). Journal of thoracic oncology : official publication of the International Association for the Study of Lung Cancer 12, 704–713 (2017); published online EpubApr (10.1016/j.jtho.2016.12.011). - DOI - PMC - PubMed
-
- Borromeo MD, Savage TK, Kollipara RK, He M, Augustyn A, Osborne JK, Girard L, Minna JD, Gazdar AF, Cobb MH, Johnson JE, ASCL1 and NEUROD1 Reveal Heterogeneity in Pulmonary Neuroendocrine Tumors and Regulate Distinct Genetic Programs. Cell reports, (2016); published online EpubJul 20 (10.1016/j.celrep.2016.06.081). - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials

