Miniaturisation of high-throughput plasmid DNA library preparation for next-generation sequencing using multifactorial optimisation
- PMID: 30723818
- PMCID: PMC6352296
- DOI: 10.1016/j.synbio.2019.01.002
Miniaturisation of high-throughput plasmid DNA library preparation for next-generation sequencing using multifactorial optimisation
Erratum in
-
Erratum regarding previously published articles.Synth Syst Biotechnol. 2020 Oct 12;5(4):328. doi: 10.1016/j.synbio.2020.10.003. eCollection 2020 Dec. Synth Syst Biotechnol. 2020. PMID: 33102826 Free PMC article.
Abstract
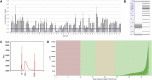
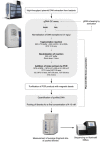
High-throughput preparation of plasmid DNA libraries for next-generation sequencing (NGS) is an important capability for molecular biology laboratories. In particular, it is an essential quality control (QC) check when large numbers of plasmid variants are being generated. Here, we describe the use of the Design of Experiments (DOE) methodology to optimise the miniaturised preparation of plasmid DNA libraries for NGS, using the Illumina® Nextera XT technology and the Labcyte Echo® acoustic liquid dispensing system. Furthermore, we describe methods which can be implemented as a QC check for identifying the presence of genomic DNA (gDNA) in plasmid DNA samples and the subsequent shearing of the gDNA, which otherwise prevents the acoustic transfer of plasmid DNA. This workflow enables the preparation of plasmid DNA libraries which yield high-quality sequencing data.
Figures







References
-
- van Dijk E.L., Auger H., Jaszczyszyn Y., Thermes C. Ten years of next-generation sequencing technology. Trends Genet. 2014;30:418–426. - PubMed
-
- Technical note: effective miniaturization of Illumina Nextera XT library prep for multiplexed whole genome sequencing and microbiome applications [internet] Labcyte Inc; San Jose, CA: 2017. http://learn.labcyte.com/rs/450-DEG-211/images/APP-G121-Miniaturization-... Available:
-
- Shapland E.B., Holmes V., Reeves C.D., Sorokin E., Durot M., Platt D. Low-cost, high-throughput sequencing of DNA assemblies using a highly multiplexed Nextera process. ACS Synth Biol. 2015;4:860–866. - PubMed
-
- Agrawal S., Cifelli S., Johnstone R., Pechter D., Barbey D.A., Lin K. Utilizing low-volume Aqueous acoustic transfer with the Echo 525 to enable miniaturization of qRT-PCR assay. J Lab Autom. 2016;21:57–63. - PubMed

