Genome-enhanced detection and identification of fungal pathogens responsible for pine and poplar rust diseases
- PMID: 30726264
- PMCID: PMC6364900
- DOI: 10.1371/journal.pone.0210952
Genome-enhanced detection and identification of fungal pathogens responsible for pine and poplar rust diseases
Abstract
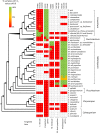
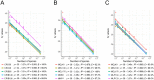
Biosurveillance is a proactive approach that may help to limit the spread of invasive fungal pathogens of trees, such as rust fungi which have caused some of the world's most damaging diseases of pines and poplars. Most of these fungi have a complex life cycle, with up to five spore stages, which is completed on two different hosts. They have a biotrophic lifestyle and may be propagated by asymptomatic plant material, complicating their detection and identification. A bioinformatics approach, based on whole genome comparison, was used to identify genome regions that are unique to the white pine blister rust fungus, Cronartium ribicola, the poplar leaf rust fungi Melampsora medusae and Melampsora larici-populina or to members of either the Cronartium and Melampsora genera. Species- and genus-specific real-time PCR assays, targeting these unique regions, were designed with the aim of detecting each of these five taxonomic groups. In total, twelve assays were developed and tested over a wide range of samples, including different spore types, different infected plant parts on the pycnio-aecial or uredinio-telial host, and captured insect vectors. One hundred percent detection accuracy was achieved for the three targeted species and two genera with either a single assay or a combination of two assays. This proof of concept experiment on pine and poplar leaf rust fungi demonstrates that the genome-enhanced detection and identification approach can be translated into effective real-time PCR assays to monitor tree fungal pathogens.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures

References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

