Genetic and transcriptomic dissection of the fiber length trait from a cotton (Gossypium hirsutum L.) MAGIC population
- PMID: 30727946
- PMCID: PMC6366115
- DOI: 10.1186/s12864-019-5427-5
Genetic and transcriptomic dissection of the fiber length trait from a cotton (Gossypium hirsutum L.) MAGIC population
Abstract
Background: Improving cotton fiber length without reducing yield is one of the major goals of cotton breeding. However, genetic improvement of cotton fiber length by breeding has been a challenge due to the narrow genetic diversity of modern cotton cultivars and negative correlations between fiber quality and yield traits. A multi-parent advanced generation inter-cross (MAGIC) population developed through random mating provides an excellent genetic resource that allows quantitative trait loci (QTL) and causal genes to be identified.
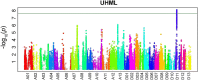
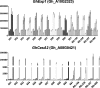
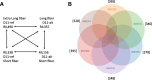
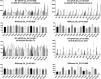
Results: An Upland cotton MAGIC population, consisting of 550 recombinant inbred lines (RILs) derived from eleven different cultivars, was used to identify fiber length QTLs and potential genes that contribute to longer fibers. A genome wide association study (GWAS) identified a cluster of single nucleotide polymorphisms (SNPs) on chromosome (Chr.) D11 that is significantly associated with fiber length. Further evaluation of the Chr. D11 genomic region among lines of the MAGIC population detected that 90% of RILs have a D11 haplotype similar to the reference TM-1 genome (D11-ref), whereas 10% of RILs inherited an alternative haplotype from one of the parents (D11-alt). The average length of fibers of D11-alt RILs was significantly shorter compared to D11-ref RILs, suggesting that alleles in the D11-alt haplotype contributed to the inferior fiber quality. RNAseq analysis of the longest and shortest fiber length RILs from D11-ref and D11-alt populations identified 949 significantly differentially expressed genes (DEGs). Gene set enrichment analysis revealed that different functional categories of genes were over-represented during fiber elongation between the four selected RILs. We found 12 genes possessing non-synonymous SNPs (nsSNPs) significantly associated with the fiber length, and three that were highly significant and were clustered at D11:24-Mb, including D11G1928, D11G1929 and D11G1931.
Conclusion: The results of this study provide insights into molecular aspects of genetic variation in fiber length and suggests candidate genes for genetic manipulation for cotton improvement.
Keywords: Cotton; Fiber length; Genome-wide association study; Multi-parent advanced generation inter cross; Quantitative trait loci; RNAseq; Single nucleotide polymorphism.
Conflict of interest statement
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Figures



References
-
- Basra AS, Malik C. Development of the cotton fiber. Int Rev Cytol. 1984;89:65–113. doi: 10.1016/S0074-7696(08)61300-5. - DOI
-
- Benedict CR, Kohel RJ, Lewis HL. Cotton fiber quality. In: Smith CW, Cothren JT, editors. Cotton: origin, history, technology, and production. New York: John Wiley & Sons; 1999;4:269–88.
-
- Zeng L, Meredith WR. Associations among lint yield, yield components, and fiber properties in an introgressed population of cotton. Crop Sci. 2009;49(5):1647–1654. doi: 10.2135/cropsci2008.09.0547. - DOI
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

