Focal adhesion kinase (FAK) activation by estrogens involves GPER in triple-negative breast cancer cells
- PMID: 30728047
- PMCID: PMC6364402
- DOI: 10.1186/s13046-019-1056-8
Focal adhesion kinase (FAK) activation by estrogens involves GPER in triple-negative breast cancer cells
Abstract
Background: Focal adhesion kinase (FAK) is a cytoplasmatic protein tyrosine kinase that associates with both integrins and growth factor receptors toward the adhesion, migration and invasion of cancer cells. The G-protein coupled estrogen receptor (GPER) has been involved in the stimulatory action of estrogens in breast tumor. In this study, we have investigated the engagement of FAK by GPER signaling in triple negative breast cancer (TNBC) cells.
Methods: Publicly available large-scale database and patient data sets derived from "The Cancer Genome Atlas" (TCGA; www.cbioportal.org ) were used to assess FAK expression in TNBC, non-TNBC tumors and normal breast tissues. MDA-MB 231 and SUM159 TNBC cells were used as model system. The levels of phosphorylated FAK, other transduction mediators and target genes were detected by western blotting analysis. Focal adhesion assay was carried out in order to determine the focal adhesion points and the formation of focal adhesions (FAs). Luciferase assays were performed to evaluate the promoters activity of c-FOS, EGR1 and CTGF upon GPER activation. The mRNA expression of the aforementioned genes was measured by real time-PCR. Boyden chamber and wound healing assays were used in order to evaluate cell migration. The statistical analysis was performed by ANOVA.
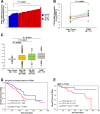
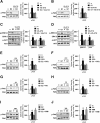
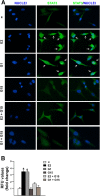
Results: We first determined by bioinformatic analysis that the mRNA expression levels of the gene encoding FAK, namely PTK2, is higher in TNBC respect to non-TNBC and normal breast tissues. Next, we found that estrogenic GPER signaling triggers Y397 FAK phosphorylation as well as the increase of focal adhesion points (FAs) in TNBC cells. Besides, we ascertained that GPER and FAK activation are involved in the STAT3 nuclear accumulation and gene expression changes. As biological counterpart, we show that FAK inhibition prevents the migration of TNBC cells upon GPER activation.
Conclusions: The present data provide novel insights regarding the action of FAK in TNBC. Moreover, on the basis of our findings estrogenic GPER signaling may be considered among the transduction mechanisms engaging FAK toward breast cancer progression.
Keywords: FAK; G-15; GPER; MDA-MB 231; STAT3, STA21; SUM159; TNBC; VS-4718.
Conflict of interest statement
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The Authors declare that they have no conflict of interest. N.N. is an employee of MSD K.K., a subsidiary of Merck & Co., Inc. and reports personal fees from MSD K. K outside this study.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Figures






References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

