Aberrant DNA methylation suppresses expression of estrogen receptor 1 (ESR1) in ovarian endometrioma
- PMID: 30728052
- PMCID: PMC6364435
- DOI: 10.1186/s13048-019-0489-1
Aberrant DNA methylation suppresses expression of estrogen receptor 1 (ESR1) in ovarian endometrioma
Abstract
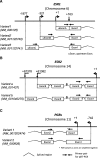
Background: In ovarian endometriomas (OE), the expression statuses of various steroid hormone receptors are altered compared with their expression statuses in eutopic endometrium (EE). For example, in OE, the expressions of estrogen receptor 1 (ESR1), which encodes ERα, and progesterone receptor (PGR) are downregulated, while the expression of ESR2, which encodes ERβ, is upregulated. The causes of these changes are unclear. DNA methylation of a specific region of a gene can result in tissue-specific gene expression. Such regions are called tissue-dependent and differentially methylated regions (T-DMRs). We previously reported that the tissue-specific expression of ESR1 is regulated by DNA methylation of a T-DMR in normal tissues. In the present study, we examined whether aberrant DNA methylation of the T-DMR is associated with the altered expressions of ESR1, ESR2 and PGR in OE.
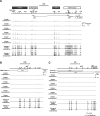
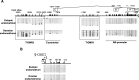
Results: Gene expression levels of ESR1, ESR2 and PGR were measured by quantitative RT-PCR. The expression levels of ESR1 and PGR were significantly lower and the expression level of ESR2 was significantly higher in OE than in EE. DNA methylation statuses were examined with an Infinium HumanMethylation450K BeadChip and sodium bisulfite sequencing. DNA methylation at the T-DMRs of ESR1 were significantly higher in OE than in EE, but no significant differences were observed in the DNA methylation statuses of ESR2 and PGR.
Conclusions: Aberrant DNA methylation of the T-DMR was associated with the impaired expression of ESR1, but not the altered expressions of ESR2 and PGR, in OE.
Keywords: DNA methylation; Endometriosis; Estrogen receptor 1; Estrogen receptor 2; Eutopic endometrium; Ovarian endometrioma; Progesterone receptor; Steroid receptor.
Conflict of interest statement
Ethics approval and consent to participate
This study was reviewed and approved by the Institutional Review Board of Yamaguchi University Graduate School of Medicine. Written informed consent was obtained from the participants before the collection of any samples, and the specimens were irreversibly de-identified. All experiments involving the handling of human tissues were performed in accordance with Tenets of the Declaration of Helsinki.
Consent for publication
Written informed consent for publication was obtained from the participants before the collection of any samples.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Figures

References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

