An integrated meta-omics approach reveals substrates involved in synergistic interactions in a bisphenol A (BPA)-degrading microbial community
- PMID: 30728080
- PMCID: PMC6366072
- DOI: 10.1186/s40168-019-0634-5
An integrated meta-omics approach reveals substrates involved in synergistic interactions in a bisphenol A (BPA)-degrading microbial community
Abstract
Background: Understanding microbial interactions in engineering bioprocesses is important to enhance and optimize performance outcomes and requires dissection of the multi-layer complexities of microbial communities. However, unraveling microbial interactions as well as substrates involved in complex microbial communities is a challenging task. Here, we demonstrate an integrated approach of metagenomics, metatranscriptomics, and targeted metabolite analysis to identify the substrates involved in interspecies interactions from a potential cross-feeding model community-bisphenol A (BPA)-biodegrading community, aiming to establish an identification method of microbial interactions in engineering or environmental bioprocesses.
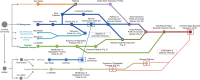
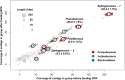
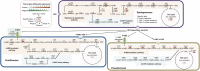
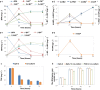
Results: The community-level BPA-metabolic pathway was constructed using integrated metagenomics and targeted metabolite analyses. The dynamics of active functions and metabolism of major community members were identified using metagenomic and metatranscriptomic analyses in concert. Correlating the community BPA biodegradation performance to the individual bacterial activities enabled the discovery of substrates involved in a synergistic interaction of cross-feeding between BPA-degrading Sphingonomas species and intermediate users, Pseudomonas sp. and Pusillimonas sp. This proposed synergistic interaction was confirmed by the co-culture of a Sphingonomas sp. and Pseudomonas sp. isolates, which demonstrated enhanced BPA biodegradation compared to the isolate of Sphingonomas sp. alone.
Conclusion: The three types of integrated meta-omics analyses effectively revealed the metabolic capability at both community-wide and individual bacterial levels. The correlation between these two levels revealed the hidden connection between apparent overall community performance and the contributions of individual community members and their interactions in a BPA-degrading microbial community. In addition, we demonstrated that using integrated multi-omics in conjunction with culture-based confirmation approach is effective to elucidate the microbial interactions affecting the performance outcome. We foresee this approach would contribute the future application and operation of environmental bioprocesses on a knowledge-based control.
Keywords: Bacterial interactions; Biodegradation; Bisphenol A; Integrated meta-omics.
Conflict of interest statement
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
Not applicable.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Figures


References
-
- Zhuang WQ, Yi S, Bill M, Brisson VL, Feng X, Men Y, Conrad ME, Tang YJ, Alvarez-Cohen L. Incomplete Wood-Ljungdahl pathway facilitates one-carbon metabolism in organohalide-respiring Dehalococcoides mccartyi. Proc Natl Acad Sci U S A. 2014;111:6419–6424. doi: 10.1073/pnas.1321542111. - DOI - PMC - PubMed
-
- Iwabuchi N, Sunairi M, Urai M, Itoh C, Anzai H, Nakajima M, Harayama S. Extracellular polysaccharides of Rhodococcus rhodochrous S-2 stimulate the degradation of aromatic components in crude oil by indigenous marine bacteria. Appl Environ Microb. 2002;68:2337–2343. doi: 10.1128/AEM.68.5.2337-2343.2002. - DOI - PMC - PubMed

