Anti-tumour immunity controlled through mRNA m6A methylation and YTHDF1 in dendritic cells
- PMID: 30728504
- PMCID: PMC6522227
- DOI: 10.1038/s41586-019-0916-x
Anti-tumour immunity controlled through mRNA m6A methylation and YTHDF1 in dendritic cells
Erratum in
-
Author Correction: Anti-tumour immunity controlled through mRNA m6A methylation and YTHDF1 in dendritic cells.Nature. 2019 Apr;568(7751):E3. doi: 10.1038/s41586-019-1046-1. Nature. 2019. PMID: 30911170
Abstract
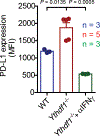
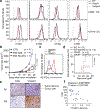
There is growing evidence that tumour neoantigens have important roles in generating spontaneous antitumour immune responses and predicting clinical responses to immunotherapies1,2. Despite the presence of numerous neoantigens in patients, complete tumour elimination is rare, owing to failures in mounting a sufficient and lasting antitumour immune response3,4. Here we show that durable neoantigen-specific immunity is regulated by mRNA N6-methyadenosine (m6A) methylation through the m6A-binding protein YTHDF15. In contrast to wild-type mice, Ythdf1-deficient mice show an elevated antigen-specific CD8+ T cell antitumour response. Loss of YTHDF1 in classical dendritic cells enhanced the cross-presentation of tumour antigens and the cross-priming of CD8+ T cells in vivo. Mechanistically, transcripts encoding lysosomal proteases are marked by m6A and recognized by YTHDF1. Binding of YTHDF1 to these transcripts increases the translation of lysosomal cathepsins in dendritic cells, and inhibition of cathepsins markedly enhances cross-presentation of wild-type dendritic cells. Furthermore, the therapeutic efficacy of PD-L1 checkpoint blockade is enhanced in Ythdf1-/- mice, implicating YTHDF1 as a potential therapeutic target in anticancer immunotherapy.
Conflict of interest statement
Figures







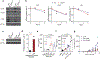



Comment in
-
Translate less, prime better, to improve anti-tumor responses.Nat Immunol. 2019 May;20(5):518-520. doi: 10.1038/s41590-019-0371-8. Nat Immunol. 2019. PMID: 30936492 No abstract available.
-
YTHDF1 Control of Dendritic Cell Cross-Priming as a Possible Target of Cancer Immunotherapy.Biochemistry. 2019 Apr 16;58(15):1945-1946. doi: 10.1021/acs.biochem.9b00200. Epub 2019 Apr 3. Biochemistry. 2019. PMID: 30943019 No abstract available.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

