Fungal Planet description sheets: 785-867
- PMID: 30728607
- PMCID: PMC6344811
- DOI: 10.3767/persoonia.2018.41.12
Fungal Planet description sheets: 785-867
Abstract
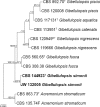
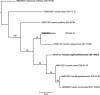
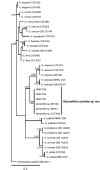
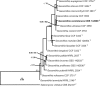
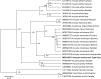
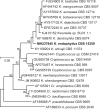
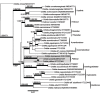
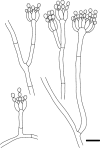
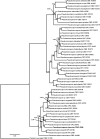
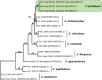
Novel species of fungi described in this study include those from various countries as follows: Angola, Gnomoniopsis angolensis and Pseudopithomyces angolensis on unknown host plants. Australia, Dothiora corymbiae on Corymbia citriodora, Neoeucasphaeria eucalypti (incl. Neoeucasphaeria gen. nov.) on Eucalyptus sp., Fumagopsis stellae on Eucalyptus sp., Fusculina eucalyptorum (incl. Fusculinaceae fam. nov.) on Eucalyptus socialis, Harknessia corymbiicola on Corymbia maculata, Neocelosporium eucalypti (incl. Neocelosporium gen. nov., Neocelosporiaceae fam. nov. and Neocelosporiales ord. nov.) on Eucalyptus cyanophylla, Neophaeomoniella corymbiae on Corymbia citriodora, Neophaeomoniella eucalyptigena on Eucalyptus pilularis, Pseudoplagiostoma corymbiicola on Corymbia citriodora, Teratosphaeria gracilis on Eucalyptus gracilis, Zasmidium corymbiae on Corymbia citriodora. Brazil, Calonectria hemileiae on pustules of Hemileia vastatrix formed on leaves of Coffea arabica, Calvatia caatinguensis on soil, Cercospora solani-betacei on Solanum betaceum, Clathrus natalensis on soil, Diaporthe poincianellae on Poincianella pyramidalis, Geastrum piquiriunense on soil, Geosmithia carolliae on wing of Carollia perspicillata, Henningsia resupinata on wood, Penicillium guaibinense from soil, Periconia caespitosa from leaf litter, Pseudocercospora styracina on Styrax sp., Simplicillium filiforme as endophyte from Citrullus lanatus, Thozetella pindobacuensis on leaf litter, Xenosonderhenia coussapoae on Coussapoa floccosa. Canary Islands (Spain), Orbilia amarilla on Euphorbia canariensis. Cape Verde Islands, Xylodon jacobaeus on Eucalyptus camaldulensis. Chile, Colletotrichum arboricola on Fuchsia magellanica. Costa Rica, Lasiosphaeria miniovina on tree branch. Ecuador, Ganoderma chocoense on tree trunk. France, Neofitzroyomyces nerii (incl. Neofitzroyomyces gen. nov.) on Nerium oleander. Ghana, Castanediella tereticornis on Eucalyptus tereticornis, Falcocladium africanum on Eucalyptus brassiana, Rachicladosporium corymbiae on Corymbia citriodora. Hungary, Entoloma silvae-frondosae in Carpinus betulus-Pinus sylvestris mixed forest. Iran, Pseudopyricularia persiana on Cyperus sp. Italy, Inocybe roseascens on soil in mixed forest. Laos, Ophiocordyceps houaynhangensis on Coleoptera larva. Malaysia, Monilochaetes melastomae on Melastoma sp. Mexico, Absidia terrestris from soil. Netherlands, Acaulium pannemaniae, Conioscypha boutwelliae, Fusicolla septimanifiniscientiae, Gibellulopsis simonii, Lasionectria hilhorstii, Lectera nordwiniana, Leptodiscella rintelii, Parasarocladium debruynii and Sarocladium dejongiae (incl. Sarocladiaceae fam. nov.) from soil. New Zealand, Gnomoniopsis rosae on Rosa sp. and Neodevriesia metrosideri on Metrosideros sp. Puerto Rico, Neodevriesia coccolobae on Coccoloba uvifera, Neodevriesia tabebuiae and Alfaria tabebuiae on Tabebuia chrysantha. Russia, Amanita paludosa on bogged soil in mixed deciduous forest, Entoloma tiliae in forest of Tilia × europaea, Kwoniella endophytica on Pyrus communis. South Africa, Coniella diospyri on Diospyros mespiliformis, Neomelanconiella combreti (incl. Neomelanconiellaceae fam. nov. and Neomelanconiella gen. nov.) on Combretum sp., Polyphialoseptoria natalensis on unidentified plant host, Pseudorobillarda bolusanthi on Bolusanthus speciosus, Thelonectria pelargonii on Pelargonium sp. Spain, Vermiculariopsiella lauracearum and Anungitopsis lauri on Laurus novocanariensis, Geosmithia xerotolerans from a darkened wall of a house, Pseudopenidiella gallaica on leaf litter. Thailand, Corynespora thailandica on wood, Lareunionomyces loeiensis on leaf litter, Neocochlearomyces chromolaenae (incl. Neocochlearomyces gen. nov.) on Chromolaena odorata, Neomyrmecridium septatum (incl. Neomyrmecridium gen. nov.), Pararamichloridium caricicola on Carex sp., Xenodactylaria thailandica (incl. Xenodactylariaceae fam. nov. and Xenodactylaria gen. nov.), Neomyrmecridium asiaticum and Cymostachys thailandica from unidentified vine. USA, Carolinigaster bonitoi (incl. Carolinigaster gen. nov.) from soil, Penicillium fortuitum from house dust, Phaeotheca shathenatiana (incl. Phaeothecaceae fam. nov.) from twig and cone litter, Pythium wohlseniorum from stream water, Superstratomyces tardicrescens from human eye, Talaromyces iowaense from office air. Vietnam, Fistulinella olivaceoalba on soil. Morphological and culture characteristics along with DNA barcodes are provided.
Keywords: ITS nrDNA barcodes; LSU; new taxa; systematics.
Figures












































































































References
-
- Abarca GH, Castañeda-Ruiz RF, Arias-Mota RA, et al. 2011. A new species of Heliocephala from México with an assessment of the systematic positions of the anamorph genera Heliocephala and Holubovaniella. Mycologia 103:631–640. - PubMed
-
- Alessio CL, Rebaudengo E. 1980. Inocybe, Iconographia Mycologica 29, Suppl. 3, Bd. 1 (Generalia et Descriptiones), Bd. 2 (Tabulae), Trento.
-
- Alfredo DS, Sousa JO, Souza EJ, et al. 2016. Novelties of gasteroid fungi, earthstars and puffballs, from the Brazilian Atlantic rainforest. In: de Madrid Anales del Jardín Botánico, Vol. 73 (2). Consejo Superior de Investigaciones Científicas.
-
- Alpago Novello L. 2006. Funghi rari e poco conosciuti della sinistra Piave in Valbelluna. Tipografia Milani, Verona.
LinkOut - more resources
Full Text Sources
