Genetic control of compound leaf development in the mungbean (Vigna radiata L.)
- PMID: 30729013
- PMCID: PMC6355865
- DOI: 10.1038/s41438-018-0088-0
Genetic control of compound leaf development in the mungbean (Vigna radiata L.)
Abstract
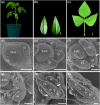
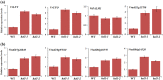
Many studies suggest that there are distinct regulatory processes controlling compound leaf development in different clades of legumes. Loss of function of the LEAFY (LFY) orthologs results in a reduction of leaf complexity to different degrees in inverted repeat-lacking clade (IRLC) and non-IRLC species. To further understand the role of LFY orthologs and the molecular mechanism in compound leaf development in non-IRLC plants, we studied leaf development in unifoliate leaf (un) mutant, a classical mutant of mungbean (Vigna radiata L.), which showed a complete conversion of compound leaves into simple leaves. Our analysis revealed that UN encoded the mungbean LFY ortholog (VrLFY) and played a significant role in leaf development. In situ RNA hybridization results showed that STM-like KNOXI genes were expressed in compound leaf primordia in mungbean. Furthermore, increased leaflet number in heptafoliate leaflets1 (hel1) mutants was demonstrated to depend on the function of VrLFY and KNOXI genes in mungbean. Our results suggested that HEL1 is a key factor coordinating distinct processes in the control of compound leaf development in mungbean and its related non-IRLC legumes.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures




References
-
- Jackson D, Veit B, Hake S. Expression of maize KNOTTED 1 related homeobox genes in the shoot apical meristem predicts patterns of morphogenesis in the vegetative shoot. Development. 1994;120:405–413.

