Distinct pathological signatures in human cellular models of myotonic dystrophy subtypes
- PMID: 30730308
- PMCID: PMC6482996
- DOI: 10.1172/jci.insight.122686
Distinct pathological signatures in human cellular models of myotonic dystrophy subtypes
Abstract
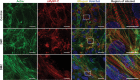
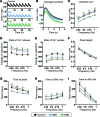
Myotonic dystrophy (DM) is the most common autosomal dominant muscular dystrophy and encompasses both skeletal muscle and cardiac complications. DM is nucleotide repeat expansion disorder in which type 1 (DM1) is due to a trinucleotide repeat expansion on chromosome 19 and type 2 (DM2) arises from a tetranucleotide repeat expansion on chromosome 3. Developing representative models of DM in animals has been challenging due to instability of nucleotide repeat expansions, especially for DM2, which is characterized by nucleotide repeat expansions often greater than 5,000 copies. To investigate mechanisms of human DM, we generated cellular models of DM1 and DM2. We used regulated MyoD expression to reprogram urine-derived cells into myotubes. In this myogenic cell model, we found impaired dystrophin expression, in the presence of muscleblind-like 1 (MBNL1) foci, and aberrant splicing in DM1 but not in DM2 cells. We generated induced pluripotent stem cells (iPSC) from healthy controls and DM1 and DM2 subjects, and we differentiated these into cardiomyocytes. DM1 and DM2 cells displayed an increase in RNA foci concomitant with cellular differentiation. iPSC-derived cardiomyocytes from DM1 but not DM2 had aberrant splicing of known target genes and MBNL sequestration. High-resolution imaging revealed tight association between MBNL clusters and RNA foci in DM1. Ca2+ transients differed between DM1- and DM2 iPSC-derived cardiomyocytes, and each differed from healthy control cells. RNA-sequencing from DM1- and DM2 iPSC-derived cardiomyocytes revealed distinct misregulation of gene expression, as well as differential aberrant splicing patterns. Together, these data support that DM1 and DM2, despite some shared clinical and molecular features, have distinct pathological signatures.
Keywords: Cardiology; Genetic variation; Muscle; Stem cells; iPS cells.
Conflict of interest statement
Figures








References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

