Normalization strategies differently affect circulating miRNA profile associated with the training status
- PMID: 30733582
- PMCID: PMC6367481
- DOI: 10.1038/s41598-019-38505-x
Normalization strategies differently affect circulating miRNA profile associated with the training status
Abstract
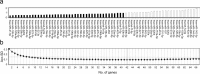
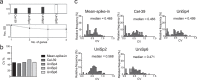
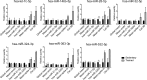
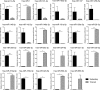
MicroRNAs are fine regulators of the whole-body adaptive response but their use as biomarkers is limited by the lack of standardized pre- and post-analytical procedures. This work aimed to compare different normalization approaches for RT-qPCR data analyses, in order to identify the most reliable and reproducible method to analyze circulating miRNA expression profiles in sedentary and highly-trained subjects. As the physically active status is known to affect miRNA expression, they could be effective biomarkers of the homeostatic response. Following RNA extraction from plasma, a panel of 179 miRNAs was assayed by RT-qPCR and quantified by applying different normalization strategies based on endogenous miRNAs and exogenous oligonucleotides. hsa-miR-320d was found as the most appropriate reference miRNA in reducing the technical variability among the experimental replicates and, hence, in highlighting the inter-cohorts differences. Our data showed an association between the physically active status and specific skeletal muscle- and bone-associated circulating miRNAs profiles, revealing that established epigenetic modifications affect the baseline physiological status of these tissues. Since different normalization strategies led to different outputs, in order to avoid misleading interpretation of data, we remark the importance of the accurate choice of the most reliable normalization method in every experimental setting.
Conflict of interest statement
The authors declare no competing interests.
Figures

References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

