A Microglial Signature Directing Human Aging and Neurodegeneration-Related Gene Networks
- PMID: 30733664
- PMCID: PMC6353788
- DOI: 10.3389/fnins.2019.00002
A Microglial Signature Directing Human Aging and Neurodegeneration-Related Gene Networks
Abstract
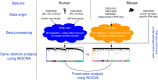
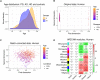
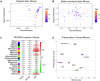
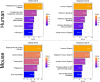
Aging is regarded as a major risk factor for neurodegenerative diseases. Thus, a better understanding of the similarities between the aging process and neurodegenerative diseases at the cellular and molecular level may reveal better understanding of this detrimental relationship. In the present study, we mined publicly available gene expression datasets from healthy individuals and patients affected by neurodegenerative diseases (Alzheimer's disease, Parkinson's disease, and Huntington's disease) across a broad age spectrum and compared those with mouse aging and mouse cell-type specific gene expression profiles. We performed weighted gene co-expression network analysis (WGCNA) and found a gene network strongly related with both aging and neurodegenerative diseases. This network was significantly enriched with a microglial signature as imputed from cell type-specific sequencing data. Since mouse models are extensively used for the study of human diseases, we further compared these human gene regulatory networks with age-specific mouse brain transcriptomes. We discovered significantly preserved networks with both human aging and human disease and identified 17 shared genes in the top-ranked immune/microglia module, among which we found five human hub genes TYROBP, FCER1G, ITGB2, MYO1F, PTPRC, and two mouse hub genes Trem2 and C1qa. Taken together, these results support the hypothesis that microglia are key players involved in human aging and neurodegenerative diseases, and suggest that mouse models should be appropriate for studying these microglial changes in human.
Keywords: Alzheimer; Parkinson; WGCNA; aging; bioinformatics; gene networks; microglia; neurodegeneration.
Figures

References
-
- Aaronson J., Rosinski J. (2015). Transcriptome profiling in knock-in mouse models of Huntington’s disease [Hippocampus_mRNA]. Available at: https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE73503
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

