Developing a High-Throughput SNP-Based Marker System to Facilitate the Introgression of Traits From Aegilops Species Into Bread Wheat (Triticum aestivum)
- PMID: 30733728
- PMCID: PMC6354564
- DOI: 10.3389/fpls.2018.01993
Developing a High-Throughput SNP-Based Marker System to Facilitate the Introgression of Traits From Aegilops Species Into Bread Wheat (Triticum aestivum)
Abstract
The genus Aegilops contains a diverse collection of wild species exhibiting variation in geographical distribution, ecological adaptation, ploidy and genome organization. Aegilops is the most closely related genus to Triticum which includes cultivated wheat, a globally important crop that has a limited gene pool for modern breeding. Aegilops species are a potential future resource for wheat breeding for traits, such as adaptation to different ecological conditions and pest and disease resistance. This study describes the development and application of the first high-throughput genotyping platform specifically designed for screening wheat relative species. The platform was used to screen multiple accessions representing all species in the genus Aegilops. Firstly, the data was demonstrated to be useful for screening diversity and examining relationships within and between Aegilops species. Secondly, markers able to characterize and track introgressions from Aegilops species in hexaploid wheat were identified and validated using two different approaches.
Keywords: Aegilops; genotyping array; introgression; single nucleotide polymorphism (SNP); wheat; wheat relative.
Figures
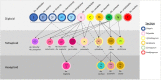



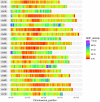

References
-
- Allen A. M., Winfield M. O., Burridge A. J., Downie R. C., Benbow H. R., Barker G. L., et al. (2016). Characterisation of a wheat breeders' array suitable for high throughput SNP genotyping of global accessions of hexaploid bread wheat (Triticum aestivium). Plant Biotechnol. J. 15, 390–401. 10.1111/pbi.12635 - DOI - PMC - PubMed
-
- Badaeva E. D., Amosova A. V., Samatadze T. E., Zoshchuk S. A., Shostak N. G., Chikida N. N., et al. (2004). Genome differentiation in aegilops. 4. evolution of the U-genome cluster. plant Syst. Evol. 246, 45–76 10.1007/s00606-003-0072-4 - DOI
-
- Burridge A. J., Wilkinson P. A., Winfield M. O., Barker G. L. A., Allen A. M., Coghill J. A., et al. (2017). Conversion of array-based single nucleotide polymorphic markers for use in targeted genotyping by sequencing in hexaploid wheat (Triticum aestivum). Plant Biotechnol. J. 16, 867–876. 10.1111/pbi.12834 - DOI - PMC - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources

