The organization of leukotriene biosynthesis on the nuclear envelope revealed by single molecule localization microscopy and computational analyses
- PMID: 30735559
- PMCID: PMC6368329
- DOI: 10.1371/journal.pone.0211943
The organization of leukotriene biosynthesis on the nuclear envelope revealed by single molecule localization microscopy and computational analyses
Abstract
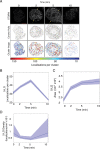
The initial steps in the synthesis of leukotrienes are the translocation of 5-lipoxygenase (5-LO) to the nuclear envelope and its subsequent association with its scaffold protein 5-lipoxygenase-activating protein (FLAP). A major gap in our understanding of this process is the knowledge of how the organization of 5-LO and FLAP on the nuclear envelope regulates leukotriene synthesis. We combined single molecule localization microscopy with Clus-DoC cluster analysis, and also a novel unbiased cluster analysis to analyze changes in the relationships between 5-LO and FLAP in response to activation of RBL-2H3 cells to generate leukotriene C4. We identified the time-dependent reorganization of both 5-LO and FLAP into higher-order assemblies or clusters in response to cell activation via the IgE receptor. Clus-DoC analysis identified a subset of these clusters with a high degree of interaction between 5-LO and FLAP that specifically correlates with the time course of LTC4 synthesis, strongly suggesting their role in the initiation of leukotriene biosynthesis.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures






References
-
- Turner H, Kinet JP. Signalling through the high-affinity IgE receptor Fc epsilonRI. Nature. 1999;402(6760 Suppl):B24–30. . - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

