Stem Cell Quiescence: Dynamism, Restraint, and Cellular Idling
- PMID: 30735649
- PMCID: PMC6413865
- DOI: 10.1016/j.stem.2019.01.001
Stem Cell Quiescence: Dynamism, Restraint, and Cellular Idling
Abstract
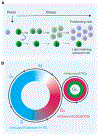
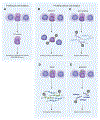
Stem cells can reside in a state of reversible growth arrest, or quiescence, for prolonged periods of time. Although quiescence has long been viewed as a dormant, low-activity state, increasing evidence suggests that quiescence represents states of poised potential and active restraint, as stem cells "idle" in anticipation of activation, proliferation, and differentiation. Improved understanding of quiescent stem cell dynamics is leading to novel approaches to enhance maintenance and repair of aged or diseased tissues. In this Review, we discuss recent advances in our understanding of stem cell quiescence and techniques enabling more refined analyses of quiescence in vivo.
Published by Elsevier Inc.
Figures


References
-
- Alvarez-Castelao B, Schanzenbächer CT, Hanus C, Glock C, Tom Dieck S, Dörrbaum AR, Bartnik I, Nassim-Assir B, Ciirdaeva E, Mueller A, et al. (2017). Cell-type-specific metabolic labeling of nascent proteomes in vivo. Nat. Biotechnol 35, 1196–1201. - PubMed
-
- Benecke BJ, Ben-Ze’ev A, and Penman S (1978). The control of mRNA production, translation and turnover in suspended and reattached anchorage-dependent fibroblasts. Cell 14, 931–939. - PubMed
-
- Bernstein BE, Mikkelsen TS, Xie X, Kamal M, Huebert DJ, Cuff J, Fry B, Meissner A, Wernig M, Plath K, et al. (2006). A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell 125, 315–326. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

