R-Loops as Cellular Regulators and Genomic Threats
- PMID: 30735654
- PMCID: PMC6402819
- DOI: 10.1016/j.molcel.2019.01.024
R-Loops as Cellular Regulators and Genomic Threats
Abstract
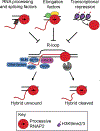
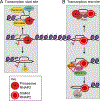
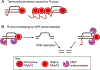
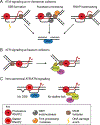
During transcription, the nascent RNA strand can base pair with its template DNA, displacing the non-template strand as ssDNA and forming a structure called an R-loop. R-loops are common across many domains of life and cause DNA damage in certain contexts. In this review, we summarize recent results implicating R-loops as important regulators of cellular processes such as transcription termination, gene regulation, and DNA repair. We also highlight recent work suggesting that R-loops can be problematic to cells as blocks to efficient transcription and replication that trigger the DNA damage response. Finally, we discuss how R-loops may contribute to cancer, neurodegeneration, and inflammatory diseases and compare the available next-generation sequencing-based approaches to map R-loops genome wide.
Copyright © 2019 Elsevier Inc. All rights reserved.
Figures


References
-
- Aguilera A, and Gomez-Gonzalez B (2017). DNA-RNA hybrids: the risks of DNA breakage during transcription. Nat Struct Mol Biol 24, 439–443. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

