Investigating the Effect of Mono- and Dimeric 360A G-Quadruplex Ligands on Telomere Stability by Single Telomere Length Analysis (STELA)
- PMID: 30736276
- PMCID: PMC6384687
- DOI: 10.3390/molecules24030577
Investigating the Effect of Mono- and Dimeric 360A G-Quadruplex Ligands on Telomere Stability by Single Telomere Length Analysis (STELA)
Abstract
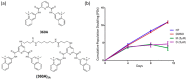
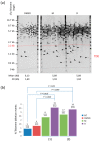
Telomeres are nucleoprotein structures that cap and protect the natural ends of chromosomes. Telomeric DNA G-rich strands can form G-quadruplex (or G4) structures. Ligands that bind to and stabilize G4 structures can lead to telomere dysfunctions by displacing shelterin proteins and/or by interfering with the replication of telomeres. We previously reported that two pyridine dicarboxamide G4 ligands, 360A and its dimeric analogue (360A)2A, were able to displace in vitro hRPA (a single-stranded DNA-binding protein of the replication machinery) from telomeric DNA by stabilizing the G4 structures. In this paper, we perform for the first time single telomere length analysis (STELA) to investigate the effect of G4 ligands on telomere length and stability. We used the unique ability of STELA to reveal the full spectrum of telomere lengths at a chromosome terminus in cancer cells treated with 360A and (360A)2A. Upon treatment with these ligands, we readily detected an increase of ultrashort telomeres, whose lengths are significantly shorter than the mean telomere length, and that could not have been detected by other methods.
Keywords: G-quadruplex ligands; G-quadruplex structures; STELA; telomere.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

