Retroelement-Linked Transcription Factor Binding Patterns Point to Quickly Developing Molecular Pathways in Human Evolution
- PMID: 30736359
- PMCID: PMC6406739
- DOI: 10.3390/cells8020130
Retroelement-Linked Transcription Factor Binding Patterns Point to Quickly Developing Molecular Pathways in Human Evolution
Erratum in
-
Correction: Nikitin, D., et al. Retroelement-Linked Transcription Factor Binding Patterns Point to Quickly Developing Molecular Pathways in Human Evolution. Cells 2019, 8, 130.Cells. 2019 Aug 5;8(8):832. doi: 10.3390/cells8080832. Cells. 2019. PMID: 31387291 Free PMC article.
Abstract
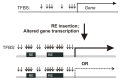
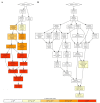
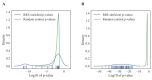
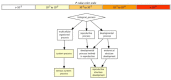
Retroelements (REs) are transposable elements occupying ~40% of the human genome that can regulate genes by providing transcription factor binding sites (TFBS). RE-linked TFBS profile can serve as a marker of gene transcriptional regulation evolution. This approach allows for interrogating the regulatory evolution of organisms with RE-rich genomes. We aimed to characterize the evolution of transcriptional regulation for human genes and molecular pathways using RE-linked TFBS accumulation as a metric. Methods: We characterized human genes and molecular pathways either enriched or deficient in RE-linked TFBS regulation. We used ENCODE database with mapped TFBS for 563 transcription factors in 13 human cell lines. For 24,389 genes and 3124 molecular pathways, we calculated the score of RE-linked TFBS regulation reflecting the regulatory evolution rate at the level of individual genes and molecular pathways. Results: The major groups enriched by RE regulation deal with gene regulation by microRNAs, olfaction, color vision, fertilization, cellular immune response, and amino acids and fatty acids metabolism and detoxication. The deficient groups were involved in translation, RNA transcription and processing, chromatin organization, and molecular signaling. Conclusion: We identified genes and molecular processes that have characteristics of especially high or low evolutionary rates at the level of RE-linked TFBS regulation in human lineage.
Keywords: ChIP-seq; Human genome evolution; gene ontology; molecular pathways; omics approach in genetics; retrotransposons; transcription factor.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures



References
-
- Lander E.S., Linton L.M., Birren B., Nusbaum C., Zody M.C., Baldwin J., Devon K., Dewar K., Doyle M., FitzHugh W., et al. Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

