An Integrative Systems Biology and Experimental Approach Identifies Convergence of Epithelial Plasticity, Metabolism, and Autophagy to Promote Chemoresistance
- PMID: 30736412
- PMCID: PMC6406733
- DOI: 10.3390/jcm8020205
An Integrative Systems Biology and Experimental Approach Identifies Convergence of Epithelial Plasticity, Metabolism, and Autophagy to Promote Chemoresistance
Abstract
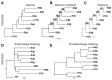
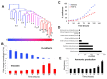
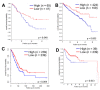
The evolution of therapeutic resistance is a major cause of death for cancer patients. The development of therapy resistance is shaped by the ecological dynamics within the tumor microenvironment and the selective pressure of the host immune system. These selective forces often lead to evolutionary convergence on pathways or hallmarks that drive progression. Thus, a deeper understanding of the evolutionary convergences that occur could reveal vulnerabilities to treat therapy-resistant cancer. To this end, we combined phylogenetic clustering, systems biology analyses, and molecular experimentation to identify convergences in gene expression data onto common signaling pathways. We applied these methods to derive new insights about the networks at play during transforming growth factor-β (TGF-β)-mediated epithelial⁻mesenchymal transition in lung cancer. Phylogenetic analyses of gene expression data from TGF-β-treated cells revealed convergence of cells toward amine metabolic pathways and autophagy during TGF-β treatment. Knockdown of the autophagy regulatory, ATG16L1, re-sensitized lung cancer cells to cancer therapies following TGF-β-induced resistance, implicating autophagy as a TGF-β-mediated chemoresistance mechanism. In addition, high ATG16L expression was found to be a poor prognostic marker in multiple cancer types. These analyses reveal the usefulness of combining evolutionary and systems biology methods with experimental validation to illuminate new therapeutic vulnerabilities for cancer.
Keywords: autophagy; epithelial–mesenchymal transition; evolution; lung cancer; metabolism; systems biology; tumor invasiveness.
Conflict of interest statement
The authors declare no conflict of interest.
Figures




References
LinkOut - more resources
Full Text Sources

