Solid-state NMR reveals a comprehensive view of the dynamics of the flexible, disordered N-terminal domain of amyloid-β fibrils
- PMID: 30737281
- PMCID: PMC6463690
- DOI: 10.1074/jbc.RA118.006559
Solid-state NMR reveals a comprehensive view of the dynamics of the flexible, disordered N-terminal domain of amyloid-β fibrils
Abstract
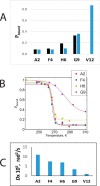
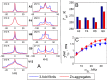
Amyloid fibril deposits observed in Alzheimer's disease comprise amyloid-β (Aβ) protein possessing a structured hydrophobic core and a disordered N-terminal domain (residues 1-16). The internal flexibility of the disordered domain is likely essential for Aβ aggregation. Here, we used 2H static solid-state NMR methods to probe the dynamics of selected side chains of the N-terminal domain of Aβ1-40 fibrils. Line shape and relaxation data suggested a two-state model in which the domain's free state undergoes a diffusive motion that is quenched in the bound state, likely because of transient interactions with the structured C-terminal domain. At 37 °C, we observed freezing of the dynamics progressively along the Aβ sequence, with the fraction of the bound state increasing and the rate of diffusion decreasing. We also found that without solvation, the diffusive motion is quenched. The solvent acted as a plasticizer reminiscent of its role in the onset of global dynamics in globular proteins. As the temperature was lowered, the fraction of the bound state exhibited sigmoidal behavior. The midpoint of the freezing curve coincided with the bulk solvent freezing for the N-terminal residues and increased further along the sequence. Using 2H R1ρ measurements, we determined the conformational exchange rate constant between the free and bound states under physiological conditions. Zinc-induced aggregation leads to the enhancement of the dynamics, manifested by the faster conformational exchange, faster diffusion, and lower freezing-curve midpoints.
Keywords: R1ρ relaxation; amyloid; fibril; intrinsically disordered protein; neurodegeneration; nuclear magnetic resonance (NMR); protein dynamics; protein misfolding; solid state NMR; zinc.
© 2019 Au et al.
Conflict of interest statement
The authors declare that they have no conflicts of interest with the contents of this article
Figures
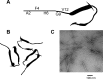
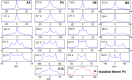
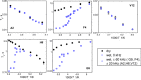
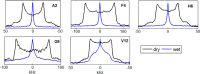
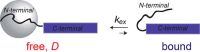

References
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

