CIC protein instability contributes to tumorigenesis in glioblastoma
- PMID: 30737375
- PMCID: PMC6368580
- DOI: 10.1038/s41467-018-08087-9
CIC protein instability contributes to tumorigenesis in glioblastoma
Abstract
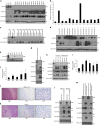
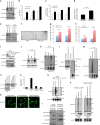
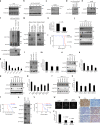
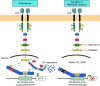
Capicua (CIC) is a transcriptional repressor that counteracts activation of genes downstream of receptor tyrosine kinase (RTK)/Ras/ERK signaling. It is well-established that tumorigenesis, especially in glioblastoma (GBM), is attributed to hyperactive RTK/Ras/ERK signaling. While CIC is mutated in other tumors, here we show that CIC has a tumor suppressive function in GBM through an alternative mechanism. We find that CIC protein levels are negligible in GBM due to continuous proteasome-mediated degradation, which is mediated by the E3 ligase PJA1 and show that this occurs through binding of CIC to its DNA target and phosphorylation on residue S173. PJA1 knockdown increased CIC stability and extended survival using in-vivo models of GBM. Deletion of the ERK binding site resulted in stabilization of CIC and increased therapeutic efficacy of ERK inhibition in GBM models. Our results provide a rationale to target CIC degradation in Ras/ERK-driven tumors, including GBM, to increase efficacy of ERK inhibitors.
Conflict of interest statement
The authors declare no competing interests.
Figures



References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

