Lymphocyte innateness defined by transcriptional states reflects a balance between proliferation and effector functions
- PMID: 30737409
- PMCID: PMC6368609
- DOI: 10.1038/s41467-019-08604-4
Lymphocyte innateness defined by transcriptional states reflects a balance between proliferation and effector functions
Abstract
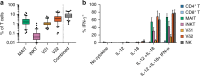
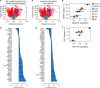
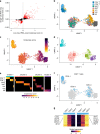
How innate T cells (ITC), including invariant natural killer T (iNKT) cells, mucosal-associated invariant T (MAIT) cells, and γδ T cells, maintain a poised effector state has been unclear. Here we address this question using low-input and single-cell RNA-seq of human lymphocyte populations. Unbiased transcriptomic analyses uncover a continuous 'innateness gradient', with adaptive T cells at one end, followed by MAIT, iNKT, γδ T and natural killer cells at the other end. Single-cell RNA-seq reveals four broad states of innateness, and heterogeneity within canonical innate and adaptive populations. Transcriptional and functional data show that innateness is characterized by pre-formed mRNA encoding effector functions, but impaired proliferation marked by decreased baseline expression of ribosomal genes. Together, our data shed new light on the poised state of ITC, in which innateness is defined by a transcriptionally-orchestrated trade-off between rapid cell growth and rapid effector function.
Conflict of interest statement
The authors declare no competing interests.
Figures





References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

