Excessive Cell Growth Causes Cytoplasm Dilution And Contributes to Senescence
- PMID: 30739799
- PMCID: PMC6386581
- DOI: 10.1016/j.cell.2019.01.018
Excessive Cell Growth Causes Cytoplasm Dilution And Contributes to Senescence
Abstract
Cell size varies greatly between cell types, yet within a specific cell type and growth condition, cell size is narrowly distributed. Why maintenance of a cell-type specific cell size is important remains poorly understood. Here we show that growing budding yeast and primary mammalian cells beyond a certain size impairs gene induction, cell-cycle progression, and cell signaling. These defects are due to the inability of large cells to scale nucleic acid and protein biosynthesis in accordance with cell volume increase, which effectively leads to cytoplasm dilution. We further show that loss of scaling beyond a certain critical size is due to DNA becoming limiting. Based on the observation that senescent cells are large and exhibit many of the phenotypes of large cells, we propose that the range of DNA:cytoplasm ratio that supports optimal cell function is limited and that ratios outside these bounds contribute to aging.
Copyright © 2019 The Author(s). Published by Elsevier Inc. All rights reserved.
Figures


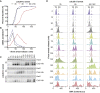


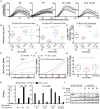

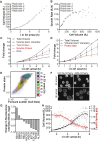


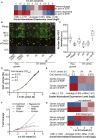


Comment in
-
Cell Biology: The Health Hazards of Super-Sizing.Curr Biol. 2019 Apr 22;29(8):R289-R292. doi: 10.1016/j.cub.2019.03.015. Curr Biol. 2019. PMID: 31014488
-
DNA Content, Cell Size, and Cell Senescence.Trends Biochem Sci. 2019 Aug;44(8):645-647. doi: 10.1016/j.tibs.2019.04.013. Epub 2019 May 31. Trends Biochem Sci. 2019. PMID: 31160123
References
Publication types
MeSH terms
Grants and funding
- R01 HD085866/HD/NICHD NIH HHS/United States
- 110275/Z/15/Z/WT_/Wellcome Trust/United Kingdom
- HHMI/Howard Hughes Medical Institute/United States
- R37 CA240765/CA/NCI NIH HHS/United States
- K01 DK098285/DK/NIDDK NIH HHS/United States
- R01 AG011085/AG/NIA NIH HHS/United States
- FC001203/WT_/Wellcome Trust/United Kingdom
- U54 CA217377/CA/NCI NIH HHS/United States
- R01 GM132447/GM/NIGMS NIH HHS/United States
- T32 GM007287/GM/NIGMS NIH HHS/United States
- FC001203/MRC_/Medical Research Council/United Kingdom
- FC001203/CRUK_/Cancer Research UK/United Kingdom
- R01 GM046498/GM/NIGMS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

