Unraveling the Developmental and Genetic Mechanisms Underpinning Floral Architecture in Proteaceae
- PMID: 30740117
- PMCID: PMC6357683
- DOI: 10.3389/fpls.2019.00018
Unraveling the Developmental and Genetic Mechanisms Underpinning Floral Architecture in Proteaceae
Abstract
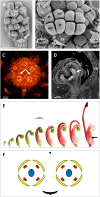
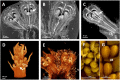
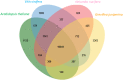
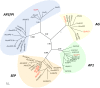
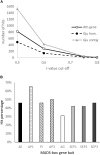
Proteaceae are a basal eudicot family with a highly conserved floral groundplan but which displays considerable variation in other aspects of floral and inflorescence morphology. Their morphological diversity and phylogenetic position make them good candidates for understanding the evolution of floral architecture, in particular the question of the homology of the undifferentiated perianth with the differentiated perianth of core eudicots, and the mechanisms underlying the repeated evolution of zygomorphy. In this paper, we combine a morphological approach to explore floral ontogenesis and a transcriptomic approach to access the genes involved in floral organ identity and development, focusing on Grevillea juniperina, a species from subfamily Grevilleoideae. We present developmental data for Grevillea juniperina and three additional species that differ in their floral symmetry using stereomicroscopy, SEM and High Resolution X-Ray Computed Tomography. We find that the adnation of stamens to tepals takes place at early developmental stages, and that the establishment of bilateral symmetry coincides with the asymmetrical growth of the single carpel. To set a framework for understanding the genetic basis of floral development in Proteaceae, we generated and annotated de novo a reference leaf/flower transcriptome from Grevillea juniperina. We found Grevillea homologs of all lineages of MADS-box genes involved in floral organ identity. Using Arabidopsis thaliana gene expression data as a reference, we found homologs of other genes involved in floral development in the transcriptome of G. juniperina. We also found at least 21 class I and class II TCP genes, a gene family involved in the regulation of growth processes, including floral symmetry. The expression patterns of a set of floral genes obtained from the transcriptome were characterized during floral development to assess their organ specificity and asymmetry of expression.
Keywords: High Resolution X-Ray Computed Tomography; MADS-box genes; Proteaceae; TCP genes; development; floral symmetry; flower; transcriptome.
Figures



References
-
- Bartlett M. E., Williams S. K., Taylor Z., DeBlasio S., Goldshmidt A., Hall D. H., et al. (2015). The maize PI/GLO ortholog Zmm16/sterile tassel silky ear1 interacts with the zygomorphy and sex determination pathways in flower development. Plant Cell 27 3081–3098. 10.1105/tpc.15.00679 - DOI - PMC - PubMed
-
- Broholm S. K., Tätiharju S., Laitinen R. A. E., Albert V. A., Heeri T. H., Elomaa P. (2008). A TCP domain transcription factor controls flower type specification along the radial axis of the Gerbera (Asteraceae) inflorescence. Proc. Nat. Acad. Sci. U.S.A. 105 9117–9122. 10.1073/pnas.0801359105 - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources

