Small-Molecule Inhibitors of the Proteasome's Regulatory Particle
- PMID: 30740849
- PMCID: PMC6765334
- DOI: 10.1002/cbic.201900017
Small-Molecule Inhibitors of the Proteasome's Regulatory Particle
Abstract
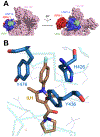
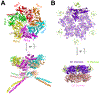
Cells need to synthesize and degrade proteins consistently. Maintaining a balanced level of protein in the cell requires a carefully controlled system and significant energy. Degradation of unwanted or damaged proteins into smaller peptide units can be accomplished by the proteasome. The proteasome is composed of two main subunits. The first is the core particle (20S CP), and within this core particle are three types of threonine proteases. The second is the regulatory complex (19S RP), which has a myriad of activities including recognizing proteins marked for degradation and shuttling the protein into the 20S CP to be degraded. Small-molecule inhibitors of the 20S CP have been developed and are exceptional treatments for multiple myeloma (MM). 20S CP inhibitors disrupt the protein balance, leading to cellular stress and eventually to cell death. Unfortunately, the 20S CP inhibitors currently available have dose-limiting off-target effects and resistance can be acquired rapidly. Herein, we discuss small molecules that have been discovered to interact with the 19S RP subunit or with a protein closely associated with 19S RP activity. These molecules still elicit their toxicity by preventing the proteasome from degrading proteins, but do so through different mechanisms of action.
Keywords: 19S RP; cancer; inhibitors; proteasome; ubiquitin.
© 2019 Wiley-VCH Verlag GmbH & Co. KGaA, Weinheim.
Figures

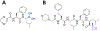




References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

