Reversible fold-switching controls the functional cycle of the antitermination factor RfaH
- PMID: 30742024
- PMCID: PMC6370827
- DOI: 10.1038/s41467-019-08567-6
Reversible fold-switching controls the functional cycle of the antitermination factor RfaH
Abstract
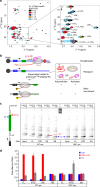
RfaH, member of the NusG/Spt5 family, activates virulence genes in Gram-negative pathogens. RfaH exists in two states, with its C-terminal domain (CTD) folded either as α-helical hairpin or β-barrel. In free RfaH, the α-helical CTD interacts with, and masks the RNA polymerase binding site on, the N-terminal domain, autoinhibiting RfaH and restricting its recruitment to opsDNA sequences. Upon activation, the domains separate and the CTD refolds into the β-barrel, which recruits a ribosome, activating translation. Using NMR spectroscopy, we show that only a complete ops-paused transcription elongation complex activates RfaH, probably via a transient encounter complex, allowing the refolded CTD to bind ribosomal protein S10. We also demonstrate that upon release from the elongation complex, the CTD transforms back into the autoinhibitory α-state, resetting the cycle. Transformation-coupled autoinhibition allows RfaH to achieve high specificity and potent activation of gene expression.
Conflict of interest statement
The authors declare no competing interests.
Figures
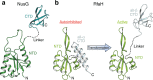

Similar articles
-
Transformation: the next level of regulation.RNA Biol. 2012 Dec;9(12):1418-23. doi: 10.4161/rna.22724. Epub 2012 Nov 6. RNA Biol. 2012. PMID: 23131843 Free PMC article.
-
An α helix to β barrel domain switch transforms the transcription factor RfaH into a translation factor.Cell. 2012 Jul 20;150(2):291-303. doi: 10.1016/j.cell.2012.05.042. Cell. 2012. PMID: 22817892 Free PMC article.
-
Flipping states: a few key residues decide the winning conformation of the only universally conserved transcription factor.Nucleic Acids Res. 2017 Sep 6;45(15):8835-8843. doi: 10.1093/nar/gkx523. Nucleic Acids Res. 2017. PMID: 28605514 Free PMC article.
-
Two Old Dogs, One New Trick: A Review of RNA Polymerase and Ribosome Interactions during Transcription-Translation Coupling.Int J Mol Sci. 2019 May 27;20(10):2595. doi: 10.3390/ijms20102595. Int J Mol Sci. 2019. PMID: 31137816 Free PMC article. Review.
-
RfaH and the ops element, components of a novel system controlling bacterial transcription elongation.Mol Microbiol. 1997 Dec;26(5):845-51. doi: 10.1046/j.1365-2958.1997.6432014.x. Mol Microbiol. 1997. PMID: 9426123 Review.
Cited by
-
RfaH Counter-Silences Inhibition of Transcript Elongation by H-NS-StpA Nucleoprotein Filaments in Pathogenic Escherichia coli.mBio. 2022 Dec 20;13(6):e0266222. doi: 10.1128/mbio.02662-22. Epub 2022 Oct 20. mBio. 2022. PMID: 36264101 Free PMC article.
-
The N-terminal domain of RfaH plays an active role in protein fold-switching.PLoS Comput Biol. 2021 Sep 3;17(9):e1008882. doi: 10.1371/journal.pcbi.1008882. eCollection 2021 Sep. PLoS Comput Biol. 2021. PMID: 34478435 Free PMC article.
-
Predicting multiple conformations via sequence clustering and AlphaFold2.Nature. 2024 Jan;625(7996):832-839. doi: 10.1038/s41586-023-06832-9. Epub 2023 Nov 13. Nature. 2024. PMID: 37956700 Free PMC article.
-
Evolutionary selection of proteins with two folds.Nat Commun. 2023 Sep 6;14(1):5478. doi: 10.1038/s41467-023-41237-2. Nat Commun. 2023. PMID: 37673981 Free PMC article.
-
Fold-switching proteins.ArXiv [Preprint]. 2025 Jul 14:arXiv:2507.10839v1. ArXiv. 2025. PMID: 40709301 Free PMC article. Updated. Preprint.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

