Mitophagy inhibits amyloid-β and tau pathology and reverses cognitive deficits in models of Alzheimer's disease
- PMID: 30742114
- PMCID: PMC6693625
- DOI: 10.1038/s41593-018-0332-9
Mitophagy inhibits amyloid-β and tau pathology and reverses cognitive deficits in models of Alzheimer's disease
Abstract
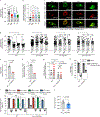
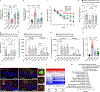
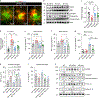
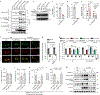
Accumulation of damaged mitochondria is a hallmark of aging and age-related neurodegeneration, including Alzheimer's disease (AD). The molecular mechanisms of impaired mitochondrial homeostasis in AD are being investigated. Here we provide evidence that mitophagy is impaired in the hippocampus of AD patients, in induced pluripotent stem cell-derived human AD neurons, and in animal AD models. In both amyloid-β (Aβ) and tau Caenorhabditis elegans models of AD, mitophagy stimulation (through NAD+ supplementation, urolithin A, and actinonin) reverses memory impairment through PINK-1 (PTEN-induced kinase-1)-, PDR-1 (Parkinson's disease-related-1; parkin)-, or DCT-1 (DAF-16/FOXO-controlled germline-tumor affecting-1)-dependent pathways. Mitophagy diminishes insoluble Aβ1-42 and Aβ1-40 and prevents cognitive impairment in an APP/PS1 mouse model through microglial phagocytosis of extracellular Aβ plaques and suppression of neuroinflammation. Mitophagy enhancement abolishes AD-related tau hyperphosphorylation in human neuronal cells and reverses memory impairment in transgenic tau nematodes and mice. Our findings suggest that impaired removal of defective mitochondria is a pivotal event in AD pathogenesis and that mitophagy represents a potential therapeutic intervention.
Figures

Comment in
-
Mitophagy and NAD+ inhibit Alzheimer disease.Autophagy. 2019 Jun;15(6):1112-1114. doi: 10.1080/15548627.2019.1596497. Epub 2019 Mar 28. Autophagy. 2019. PMID: 30922179 Free PMC article.
References
-
- Lustbader JW et al. ABAD directly links Aß to mitochondrial toxicity in Alzheimer’s disease. Science 304, 448–452 (2004). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

