Mutation in DDM1 inhibits the homology directed repair of double strand breaks
- PMID: 30742642
- PMCID: PMC6370192
- DOI: 10.1371/journal.pone.0211878
Mutation in DDM1 inhibits the homology directed repair of double strand breaks
Abstract
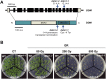
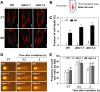
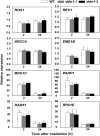
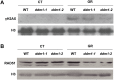
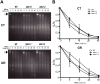
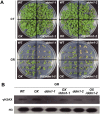
In all organisms, DNA damage must be repaired quickly and properly, as it can be lethal for cells. Because eukaryotic DNA is packaged into nucleosomes, the structural units of chromatin, chromatin modification is necessary during DNA damage repair and is achieved by histone modification and chromatin remodeling. Chromatin remodeling proteins therefore play important roles in the DNA damage response (DDR) by modifying the accessibility of DNA damage sites. Here, we show that mutation in a SWI2/SNF2 chromatin remodeling protein (DDM1) causes hypersensitivity in the DNA damage response via defects in single-strand annealing (SSA) repair of double-strand breaks (DSBs) as well as in the initial steps of homologous recombination (HR) repair. ddm1 mutants such as ddm1-1 and ddm1-2 exhibited increased root cell death and higher DSB frequency compared to the wild type after gamma irradiation. Although the DDM1 mutation did not affect the expression of most DDR genes, it did cause substantial decrease in the frequency of SSA as well as partial inhibition in the γ-H2AX and Rad51 induction, the initial steps of HR. Furthermore, global chromatin structure seemed to be affected by DDM1 mutations. These results suggest that DDM1 is involved in the homology directed repair such as SSA and HR, probably by modifying chromatin structure.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures

Similar articles
-
An insight into understanding the coupling between homologous recombination mediated DNA repair and chromatin remodeling mechanisms in plant genome: an update.Cell Cycle. 2021 Sep;20(18):1760-1784. doi: 10.1080/15384101.2021.1966584. Epub 2021 Aug 26. Cell Cycle. 2021. PMID: 34437813 Free PMC article. Review.
-
The requirement for recombination factors differs considerably between different pathways of homologous double-strand break repair in somatic plant cells.Plant J. 2012 Dec;72(5):781-90. doi: 10.1111/j.1365-313X.2012.05119.x. Epub 2012 Oct 1. Plant J. 2012. PMID: 22860689
-
ddm1 plants are sensitive to methyl methane sulfonate and NaCl stresses and are deficient in DNA repair.Plant Cell Rep. 2012 Sep;31(9):1549-61. doi: 10.1007/s00299-012-1269-1. Epub 2012 Apr 27. Plant Cell Rep. 2012. PMID: 22538524
-
Involvement of the Arabidopsis SWI2/SNF2 chromatin remodeling gene family in DNA damage response and recombination.Genetics. 2006 Jun;173(2):985-94. doi: 10.1534/genetics.105.051664. Epub 2006 Mar 17. Genetics. 2006. PMID: 16547115 Free PMC article.
-
Nucleosome remodelers in double-strand break repair.Curr Opin Genet Dev. 2013 Apr;23(2):174-84. doi: 10.1016/j.gde.2012.12.008. Epub 2013 Jan 23. Curr Opin Genet Dev. 2013. PMID: 23352131 Review.
Cited by
-
Chromatin remodeling of histone H3 variants underlies epigenetic inheritance of DNA methylation.bioRxiv [Preprint]. 2023 Aug 2:2023.07.11.548598. doi: 10.1101/2023.07.11.548598. bioRxiv. 2023. Update in: Cell. 2023 Sep 14;186(19):4100-4116.e15. doi: 10.1016/j.cell.2023.08.001. PMID: 37503143 Free PMC article. Updated. Preprint.
-
Molecular basis of chromatin remodelling by DDM1 involved in plant DNA methylation.Nat Plants. 2024 Mar;10(3):374-380. doi: 10.1038/s41477-024-01640-z. Epub 2024 Feb 27. Nat Plants. 2024. PMID: 38413824
-
Chromatin Remodeling and Epigenetic Regulation in Plant DNA Damage Repair.Int J Mol Sci. 2019 Aug 22;20(17):4093. doi: 10.3390/ijms20174093. Int J Mol Sci. 2019. PMID: 31443358 Free PMC article. Review.
-
Efficient enhancement of the antimicrobial activity of Chlamydomonas reinhardtii extract by transgene expression and molecular modification using ionizing radiation.Biotechnol Biofuels Bioprod. 2024 Oct 1;17(1):125. doi: 10.1186/s13068-024-02575-5. Biotechnol Biofuels Bioprod. 2024. PMID: 39354614 Free PMC article.
-
Seminars in cell and development biology on histone variants remodelers of H2A variants associated with heterochromatin.Semin Cell Dev Biol. 2023 Feb 15;135:93-101. doi: 10.1016/j.semcdb.2022.02.026. Epub 2022 Mar 4. Semin Cell Dev Biol. 2023. PMID: 35249811 Free PMC article. Review.
References
-
- Ravanat JL, Douki T. UV and ionizing radiations induced DNA damage, differences and similarities. Radiat Phys Chem. 2016; 128: 92–102.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials

